Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2020-06-25 , DOI: 10.1016/j.csbj.2020.06.026 Gue-Ho Hwang 1 , Jihyeon Yu 1, 2 , Soyeon Yang 1 , Woo Jae Son 1 , Kayeong Lim 3 , Heon Seok Kim 4 , Jin-Soo Kim 3, 5 , Sangsu Bae 1, 2
|
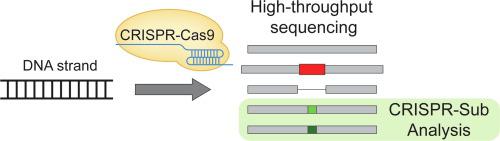
CRISPR-Cas9 induces DNA cleavages at desired target sites in a guide RNA-dependent manner; DNA editing occurs through the resulting activity of DNA repair processes including non-homologous end joining (NHEJ), which is dominant in mammalian cells. NHEJ repair frequently causes small insertions and deletions (indels) near DNA cleavage sites but only rarely causes nucleotide substitutions. High-throughput sequencing is the primary means of assessing indel and substitution frequencies in bulk populations of cells in the gene editing field. However, it is difficult to detect bona fide substitutions, which are embedded among experimentally-induced substitution errors, in high-throughput sequencing data. Here, we developed a novel analysis method, named CRISPR-Sub, to statistically detect Cas9-mediated substitutions in high-throughput sequencing data by comparing Mock- and CRISPR-treated samples. We first pinpointed ‘hotspot positions’ in target sequences at which substitution mutations were quantitatively observed much more often (p > 0.001) in CRISPR- versus Mock-treated samples. We refer to the substitution mutations in defined hotspot positions as ‘apparent substitutions’ and ultimately calculated ‘apparent substitution frequencies’ for each target. By examining 51 endogenous target sites in HeLa cells, we found that the average apparent substitution frequency was 0.8% in all queries, that apparent substitutions frequently occur near CRISPR-Cas9 cleavage sites, and that nucleotide conversion showed no meaningful nucleotide preference patterns. Furthermore, we generated NHEJ-inhibited cell lines (LIG4−/−) by knockout of the gene encoding ligase IV and found that the apparent substitution frequencies were significantly decreased in LIG4−/− cells, strongly suggesting that DNA substitutions are generated by the NHEJ pathway.
中文翻译:

CRISPR-sub:分析人细胞中CRISPR-Cas9引起的DNA取代突变。
CRISPR-Cas9以依赖RNA的方式诱导所需靶位点的DNA切割; DNA编辑是通过包括非同源末端连接(NHEJ)在内的DNA修复过程的活动而发生的,该过程在哺乳动物细胞中占主导地位。NHEJ修复经常在DNA切割位点附近引起小的插入和缺失(indels),但很少引起核苷酸取代。高通量测序是评估基因编辑领域中大量细胞内插入缺失和取代频率的主要手段。但是,在高通量测序数据中很难检测到真正的替换,这些替换嵌入在实验引起的替换错误中。在这里,我们开发了一种新颖的分析方法,名为CRISPR-Sub,通过比较经Mock和CRISPR处理的样品来统计检测高通量测序数据中Cas9介导的取代。我们首先查明了靶序列中的“热点位置”,在该序列中,在CRISPR处理过的样品中比在模拟处理过的样品中观察到替换突变的频率更高(p> 0.001)。我们将定义的热点位置中的替代突变称为“表观替代”,并最终为每个目标计算出“表观替代频率”。通过检查HeLa细胞中的51个内源性靶位点,我们发现在所有查询中平均表观取代频率为0.8%,表观取代经常发生在CRISPR-Cas9裂解位点附近,并且核苷酸转化没有显示出有意义的核苷酸偏好模式。此外,我们生成了NHEJ抑制的细胞系(我们首先查明了靶序列中的“热点位置”,在该序列中,在CRISPR处理过的样品中比在模拟处理过的样品中观察到替换突变的频率更高(p> 0.001)。我们将定义的热点位置中的替代突变称为“表观替代”,并最终为每个目标计算出“表观替代频率”。通过检查HeLa细胞中的51个内源性靶位点,我们发现在所有查询中平均表观取代频率为0.8%,表观取代经常发生在CRISPR-Cas9裂解位点附近,并且核苷酸转化没有显示出有意义的核苷酸偏好模式。此外,我们生成了NHEJ抑制的细胞系(我们首先查明了靶序列中的“热点位置”,在该序列中,在CRISPR处理过的样品中比在模拟处理过的样品中观察到替换突变的频率更高(p> 0.001)。我们将定义的热点位置中的替代突变称为“表观替代”,并最终为每个目标计算出“表观替代频率”。通过检查HeLa细胞中的51个内源性靶位点,我们发现在所有查询中平均表观取代频率为0.8%,表观取代经常发生在CRISPR-Cas9裂解位点附近,并且核苷酸转化没有显示出有意义的核苷酸偏好模式。此外,我们生成了NHEJ抑制的细胞系(001)在CRISPR和Mock处理的样本中。我们将定义的热点位置中的替代突变称为“表观替代”,并最终为每个目标计算出“表观替代频率”。通过检查HeLa细胞中的51个内源性靶位点,我们发现在所有查询中平均表观取代频率为0.8%,表观取代经常发生在CRISPR-Cas9裂解位点附近,并且核苷酸转化没有显示出有意义的核苷酸偏好模式。此外,我们生成了NHEJ抑制的细胞系(001)在CRISPR和Mock处理的样本中。我们将定义的热点位置中的替代突变称为“表观替代”,并最终为每个目标计算出“表观替代频率”。通过检查HeLa细胞中的51个内源性靶位点,我们发现在所有查询中平均表观取代频率为0.8%,表观取代经常发生在CRISPR-Cas9裂解位点附近,并且核苷酸转化没有显示出有意义的核苷酸偏好模式。此外,我们生成了NHEJ抑制的细胞系(我们发现在所有查询中平均表观取代频率为0.8%,表观取代经常发生在CRISPR-Cas9裂解位点附近,并且核苷酸转化没有显示出有意义的核苷酸偏好模式。此外,我们生成了NHEJ抑制的细胞系(我们发现在所有查询中平均表观取代频率为0.8%,表观取代经常发生在CRISPR-Cas9裂解位点附近,并且核苷酸转化没有显示出有意义的核苷酸偏好模式。此外,我们生成了NHEJ抑制的细胞系(LIG4- /)通过敲除连接酶IV的基因,发现LIG4 -/-细胞中的表观取代频率显着降低,强烈暗示DNA取代是由NHEJ途径产生的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号