Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics ( IF 2.5 ) Pub Date : 2020-06-25 , DOI: 10.1016/j.bbapap.2020.140477 Xiaoyong Pan 1 , Lin Lu 2 , Yu-Dong Cai 3
|
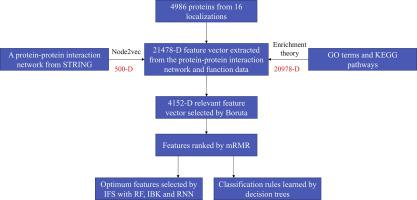
The subcellular location of a protein is highly related to its function. Identifying the location of a given protein is an essential step for investigating its related problems. Traditional experimental methods can produce solid determination. However, their limitations, such as high cost and low efficiency, are evident. Computational methods provide an alternative means to address these problems. Most previous methods constantly extract features from protein sequences or structures for building prediction models. In this study, we use two types of features and combine them to construct the model. The first feature type is extracted from a protein–protein interaction network to abstract the relationship between the encoded protein and other proteins. The second type is obtained from gene ontology and biological pathways to indicate the existing functions of the encoded protein. These features are analyzed using some feature selection methods. The final optimum features are adopted to build the model with recurrent neural network as the classification algorithm. Such model yields good performance with Matthews correlation coefficient of 0.844. A decision tree is used as a rule learning classifier to extract decision rules. Although the performance of decision rules is poor, they are valuable in revealing the molecular mechanism of proteins with different subcellular locations. The final analysis confirms the reliability of the extracted rules. The source code of the propose method is freely available at https://github.com/xypan1232/rnnloc
中文翻译:

通过网络嵌入和富集功能预测蛋白质亚细胞位置。
蛋白质的亚细胞位置与其功能高度相关。鉴定给定蛋白质的位置是研究其相关问题的重要步骤。传统的实验方法可以确定。但是,它们的局限性,例如高成本和低效率,是显而易见的。计算方法提供了解决这些问题的替代方法。以前的大多数方法都会不断地从蛋白质序列或结构中提取特征,以建立预测模型。在这项研究中,我们使用两种类型的特征并将其组合以构建模型。从蛋白质-蛋白质相互作用网络中提取第一个特征类型,以提取编码的蛋白质和其他蛋白质之间的关系。第二种是从基因本体论和生物学途径获得的,以表明编码蛋白的现有功能。使用某些特征选择方法来分析这些特征。采用最终的最优特征,以递归神经网络作为分类算法,建立了模型。这种模型在Matthews相关系数为0.844时表现出良好的性能。决策树用作规则学习分类器以提取决策规则。尽管决策规则的性能很差,但是它们对于揭示具有不同亚细胞位置的蛋白质的分子机制非常有价值。最终分析确认了提取规则的可靠性。可以在https://github.com/xypan1232/rnnloc上免费获得proposal方法的源代码。使用某些特征选择方法来分析这些特征。采用最终的最优特征,以递归神经网络作为分类算法,建立了模型。这种模型在Matthews相关系数为0.844时表现出良好的性能。决策树用作规则学习分类器以提取决策规则。尽管决策规则的性能很差,但是它们对于揭示具有不同亚细胞位置的蛋白质的分子机制非常有价值。最终分析确认了提取规则的可靠性。可以在https://github.com/xypan1232/rnnloc上免费获得proposal方法的源代码。使用某些特征选择方法来分析这些特征。采用最终的最优特征,以递归神经网络作为分类算法,建立了模型。这种模型在Matthews相关系数为0.844时表现出良好的性能。决策树用作规则学习分类器以提取决策规则。尽管决策规则的性能很差,但是它们对于揭示具有不同亚细胞位置的蛋白质的分子机制非常有价值。最终分析确认了提取规则的可靠性。可以在https://github.com/xypan1232/rnnloc上免费获得proposal方法的源代码。这种模型在Matthews相关系数为0.844时表现出良好的性能。决策树用作规则学习分类器以提取决策规则。尽管决策规则的性能很差,但是它们对于揭示具有不同亚细胞位置的蛋白质的分子机制非常有价值。最终分析确认了提取规则的可靠性。可以在https://github.com/xypan1232/rnnloc上免费获得proposal方法的源代码。这种模型在Matthews相关系数为0.844时表现出良好的性能。决策树用作规则学习分类器以提取决策规则。尽管决策规则的性能很差,但是它们对于揭示具有不同亚细胞位置的蛋白质的分子机制非常有价值。最终分析确认了提取规则的可靠性。可以在https://github.com/xypan1232/rnnloc上免费获得proposal方法的源代码。











































 京公网安备 11010802027423号
京公网安备 11010802027423号