当前位置:
X-MOL 学术
›
Macromol. Rapid Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Photoluminescent Response of Poly(3-methylthiophene)-DNA Single Nanowire Correlating to Nucleotide-Mismatch Locus in DNA-DNA Hybridization.
Macromolecular Rapid Communications ( IF 4.2 ) Pub Date : 2020-06-24 , DOI: 10.1002/marc.202000164 Dong Hyuk Park 1 , Chunzhi Cui 2 , Dong June Ahn 3, 4
Macromolecular Rapid Communications ( IF 4.2 ) Pub Date : 2020-06-24 , DOI: 10.1002/marc.202000164 Dong Hyuk Park 1 , Chunzhi Cui 2 , Dong June Ahn 3, 4
Affiliation
|
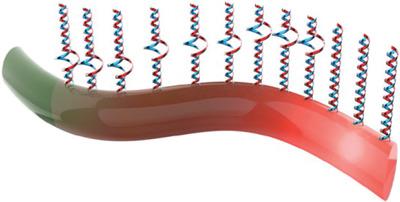
π‐Conjugated polymers have become qualified candidates for biosensing owing to their unique optoelectronic properties and excellent biocompatibility. In this contribution, nucleotide mismatches in DNA hybridization, being variable in position, are reflected in a stark manner by poly(3‐methylthiophene) (P3MT) nanowires (NWs), in which probe DNA sequence is properly functionalized. Selected as the systematic investigation are complementary target DNA (tDNA), random sequence DNA, and three kinds of 1‐mer mismatched tDNAs with different mismatch loci away from the NW's surface. Nanoscale optical observation of the single P3MT NWs in solid states reveals that the more distant the mismatch position is from the surface, the higher the photoluminescence (PL) occurs, while the complementary sequence yields the highest but the random one remains the lowest. Hence, the PL intensity increases with the relative length of the DNA–DNA hybridization from the surface. These results deliver a new basis that π‐conjugated polymers can be potentially applicable to detailed nucleotide analyses as in single nucleotide polymorphism.
中文翻译:

聚(3-甲基噻吩)-DNA单纳米线在DNA-DNA杂交中与核苷酸错配位点的光致发光反应。
π共轭聚合物由于其独特的光电性能和出色的生物相容性,已成为生物传感的合格候选物。在这一贡献中,DNA杂交中核苷酸错配的位置可变,可通过聚(3-甲基噻吩)(P3MT)纳米线(NW)以鲜明的方式反映出来,其中探针DNA序列已得到适当的功能化。被选为系统研究对象的是互补靶DNA(tDNA),随机序列DNA和三种错位位点在距西北表面不远的三种1聚体错配tDNA。单个固态P3MT NW的纳米级光学观察表明,失配位置离表面越远,发生的光致发光(PL)越高,互补序列产生最高的序列,而随机序列保持最低。因此,PL强度随表面DNA-DNA杂交的相对长度而增加。这些结果提供了一个新的基础,即π共轭聚合物可以像单核苷酸多态性一样适用于详细的核苷酸分析。
更新日期:2020-06-24
中文翻译:

聚(3-甲基噻吩)-DNA单纳米线在DNA-DNA杂交中与核苷酸错配位点的光致发光反应。
π共轭聚合物由于其独特的光电性能和出色的生物相容性,已成为生物传感的合格候选物。在这一贡献中,DNA杂交中核苷酸错配的位置可变,可通过聚(3-甲基噻吩)(P3MT)纳米线(NW)以鲜明的方式反映出来,其中探针DNA序列已得到适当的功能化。被选为系统研究对象的是互补靶DNA(tDNA),随机序列DNA和三种错位位点在距西北表面不远的三种1聚体错配tDNA。单个固态P3MT NW的纳米级光学观察表明,失配位置离表面越远,发生的光致发光(PL)越高,互补序列产生最高的序列,而随机序列保持最低。因此,PL强度随表面DNA-DNA杂交的相对长度而增加。这些结果提供了一个新的基础,即π共轭聚合物可以像单核苷酸多态性一样适用于详细的核苷酸分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号