当前位置:
X-MOL 学术
›
Ecol. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Advancing biodiversity assessments with environmental DNA: Long-read technologies help reveal the drivers of Amazonian fungal diversity.
Ecology and Evolution ( IF 2.3 ) Pub Date : 2020-06-23 , DOI: 10.1002/ece3.6477 Camila D Ritter 1, 2, 3 , Micah Dunthorn 1 , Sten Anslan 4 , Vitor Xavier de Lima 5 , Leho Tedersoo 6 , Rolf Henrik Nilsson 2, 3 , Alexandre Antonelli 2, 3, 7
Ecology and Evolution ( IF 2.3 ) Pub Date : 2020-06-23 , DOI: 10.1002/ece3.6477 Camila D Ritter 1, 2, 3 , Micah Dunthorn 1 , Sten Anslan 4 , Vitor Xavier de Lima 5 , Leho Tedersoo 6 , Rolf Henrik Nilsson 2, 3 , Alexandre Antonelli 2, 3, 7
Affiliation
|
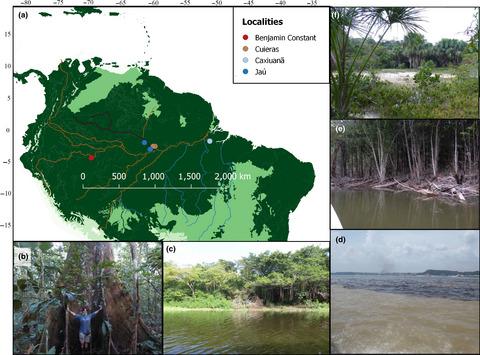
Fungi are a key component of tropical biodiversity. However, due to their inconspicuous and largely subterranean nature, they are usually neglected in biodiversity inventories. The goal of this study was to identify the key determinants of fungal richness, community composition, and turnover in tropical rainforests. We tested specifically for the effect of soil properties, habitat, and locality in Amazonia. For these analyses, we used high‐throughput sequencing data of short and long reads of fungal DNA present in soil and organic litter samples, combining existing and novel genomic data. Habitat type (phytophysiognomy) emerges as the strongest factor explaining fungal community composition. Naturally open areas—campinas—are the richest habitat overall. Soil properties have different effects depending on the soil layer (litter or mineral soil) and the choice of genetic marker. We suggest that campinas could be a neglected hotspot of fungal diversity. An underlying cause for their rich diversity may be the overall low soil fertility, which increases the reliance on biotic interactions essential for nutrient absorption in these environments, notably ectomycorrhizal fungi–plant associations. Our results highlight the advantages of using both short and long DNA reads produced through high‐throughput sequencing to characterize fungal diversity. While short reads can suffice for diversity and community comparison, long reads add taxonomic precision and have the potential to reveal population diversity.
中文翻译:

利用环境 DNA 推进生物多样性评估:长读技术有助于揭示亚马逊真菌多样性的驱动因素。
真菌是热带生物多样性的重要组成部分。然而,由于它们不显眼且大部分位于地下,因此在生物多样性清单中通常被忽视。本研究的目的是确定热带雨林真菌丰富度、群落组成和周转率的关键决定因素。我们专门测试了亚马逊流域土壤特性、栖息地和地点的影响。在这些分析中,我们使用了土壤和有机垃圾样本中真菌 DNA 的短读长和长读长的高通量测序数据,并结合了现有和新的基因组数据。栖息地类型(植物面貌)成为解释真菌群落组成的最强因素。自然开放区域——坎皮纳斯——是总体上最丰富的栖息地。根据土层(枯落物或矿质土)和遗传标记的选择,土壤特性具有不同的影响。我们认为坎皮纳斯可能是一个被忽视的真菌多样性热点地区。其丰富多样性的一个根本原因可能是整体土壤肥力较低,这增加了对这些环境中养分吸收所必需的生物相互作用的依赖,特别是外生菌根真菌与植物的关联。我们的结果强调了使用通过高通量测序产生的短和长 DNA 读数来表征真菌多样性的优势。虽然短读取足以进行多样性和群落比较,但长读取增加了分类精度,并有可能揭示种群多样性。
更新日期:2020-07-30
中文翻译:

利用环境 DNA 推进生物多样性评估:长读技术有助于揭示亚马逊真菌多样性的驱动因素。
真菌是热带生物多样性的重要组成部分。然而,由于它们不显眼且大部分位于地下,因此在生物多样性清单中通常被忽视。本研究的目的是确定热带雨林真菌丰富度、群落组成和周转率的关键决定因素。我们专门测试了亚马逊流域土壤特性、栖息地和地点的影响。在这些分析中,我们使用了土壤和有机垃圾样本中真菌 DNA 的短读长和长读长的高通量测序数据,并结合了现有和新的基因组数据。栖息地类型(植物面貌)成为解释真菌群落组成的最强因素。自然开放区域——坎皮纳斯——是总体上最丰富的栖息地。根据土层(枯落物或矿质土)和遗传标记的选择,土壤特性具有不同的影响。我们认为坎皮纳斯可能是一个被忽视的真菌多样性热点地区。其丰富多样性的一个根本原因可能是整体土壤肥力较低,这增加了对这些环境中养分吸收所必需的生物相互作用的依赖,特别是外生菌根真菌与植物的关联。我们的结果强调了使用通过高通量测序产生的短和长 DNA 读数来表征真菌多样性的优势。虽然短读取足以进行多样性和群落比较,但长读取增加了分类精度,并有可能揭示种群多样性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号