Journal of Molecular Graphics and Modelling ( IF 2.7 ) Pub Date : 2020-06-24 , DOI: 10.1016/j.jmgm.2020.107657 Adam Jo J Elatico 1 , Ricky B Nellas 1
|
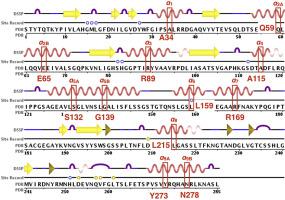
Lipases are important enzymes in many biochemical industries, thus making them attractive targets for protein engineering to improve enzymatic properties. In this work, a ‘‘reverse engineering’’ approach was explored: disrupt secondary structures to determine their contribution to enzyme stability and activity. All the α-helices of the lipase from Pseudomonas aeruginosa PAO1 (PAL) were systematically disrupted using computational proline mutagenesis and molecular dynamics (MD) simulations. This method identified the mutant (R89P), located within the vicinity of the active site, to be significantly important for stability and activity. In addition, the system (L159P), part of the ‘‘cap’’ domain that regulates substrate entry into the active site, was found to be critical for activity as it pushed the lipase to adopt a completely closed conformation. The perturbation introduced by the proline mutations resulted in increased backbone flexibility that significantly decreased protein stability. Moreover, mutations within the cap domain helices — (A115P), (S132P, G139P), (L159P), and (R169P) — resulted in increased flexibility of the N-terminal region of the helix, the mobile ‘‘lid’’ helix, that pushes the gorge into a partially closed conformation. The mutation (L159P) further increased the flexibility of the helix–loop region at the C-terminal end of the helix to push the lid into the fully closed state. Therefore, the and helices could be ‘‘hot spots’’ for stabilizing mutations that could improve the overall enzyme stability and activity this lipase. The insights obtained in this work may be validated experimentally in future works.
中文翻译:

铜绿假单胞菌PAO1中脂肪酶的计算逆向工程:α-螺旋。
脂肪酶在许多生化工业中是重要的酶,因此使其成为蛋白质工程以提高酶学性质的有吸引力的靶标。在这项工作中,探索了一种“逆向工程”方法:破坏二级结构,以确定其对酶稳定性和活性的贡献。铜绿假单胞菌PAO1(PAL)的脂肪酶的所有α螺旋都使用计算脯氨酸诱变和分子动力学(MD)模拟系统地破坏了。该方法确定了突变(R89P),位于活性位点附近,对于稳定性和活性非常重要。除此之外系统(L159P)是调节底物进入活性位点的“帽”结构域的一部分,被发现对于活性至关重要,因为它推动脂肪酶采取完全封闭的构象。脯氨酸突变引起的扰动导致主链柔性增加,从而大大降低了蛋白质的稳定性。此外,帽结构域螺旋内的突变- (A115P), (S132P,G139P), (L159P),以及 (R169P)-增强了N末端区域的柔韧性 螺旋,移动的“盖”螺旋,将峡谷推向部分封闭的状态。的 突变(L159P)进一步增加了C末端螺旋环区域的灵活性 螺旋将盖推入完全关闭状态。因此, 和 螺旋可能是稳定突变的“热点”,可以改善整体酶的稳定性和这种脂肪酶的活性。这项工作中获得的见识可以在未来的工作中通过实验进行验证。











































 京公网安备 11010802027423号
京公网安备 11010802027423号