Current Pharmaceutical Biotechnology ( IF 2.2 ) Pub Date : 2021-02-28 , DOI: 10.2174/1389201021666200610105549 Kang M Lee 1 , Cheorl-Ho Kim 2 , Jong H Kim 1 , Sung S Kim 1 , Seung-Hak Cho 1
|
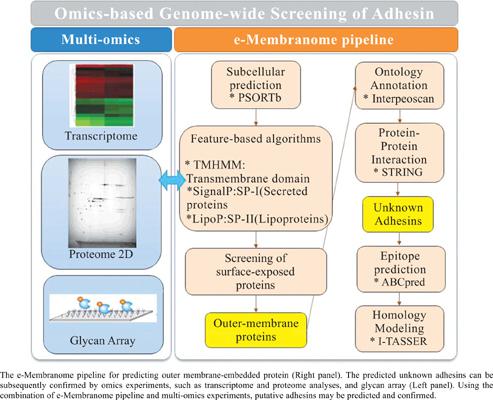
Objective: Lectin-like adhesins of enteric bacterial pathogens such as Escherichia coli are an attractive target for vaccine or drug development. Here, we have developed e-Membranome as a database of genome-wide putative adhesins in Escherichia coli (E. coli).
Methods: The outer membrane adhesins were predicted from the annotated genes of Escherichia coli strains using the PSORTb program. Further analysis was performed using Interproscan and the String database. The candidate proteins can be investigated for homology modeling of the Three-Dimensional (3D) structure (I-TASSER version 5.1), epitope region (ABCpred), and the glycan array.
Results: e-Membranome is implemented using the Django (version 2.2.5) framework. The Web Application Server Apache Tomcat 6.0 is integrated into the platform on Ubuntu Linux (version 16.04). MySQL database (version 5.7) is used as a database engine. The information on homology model of the 3D structure, epitope region, and affinity information from the glycan array will be stored in the e- Membranome database. As a case study, we performed a genome-wide screening of outer membraneembedded proteins from the annotated genes of E. coli using the e-Membranome pipeline.
Conclusion: This platform is expected to be a valuable resource for advancing research of outer membrane proteins for the construction of lectin-glycan interaction network of E. coli. In addition, the e- Membranome pipeline can be extended to other similar biological systems that need to address hostpathogen interactions.
中文翻译:

电子膜组:大肠埃希菌外膜蛋白的全基因组分析数据库
目的:肠细菌病原体(例如大肠杆菌)的凝集素样粘附素是疫苗或药物开发的有吸引力的靶标。在这里,我们开发了e-膜组作为大肠杆菌(E. coli)中全基因组推定粘附素的数据库。
方法:使用PSORTb程序从大肠杆菌菌株的注释基因中预测外膜粘附素。使用Interproscan和String数据库进行了进一步的分析。可以研究候选蛋白质,以进行三维(3D)结构(I-TASSER版本5.1),表位区域(ABCpred)和聚糖阵列的同源性建模。
结果:电子膜是使用Django(2.2.5版)框架实现的。Web应用程序服务器Apache Tomcat 6.0已集成到Ubuntu Linux(版本16.04)上的平台中。MySQL数据库(5.7版)用作数据库引擎。有关3D结构,表位区域的同源性模型的信息以及来自聚糖阵列的亲和力信息将存储在e-膜组数据库中。作为案例研究,我们使用e-膜组管线从大肠杆菌的带注释基因中对外膜嵌入蛋白进行了全基因组筛选。
结论:该平台有望为推进外膜蛋白的研究提供有价值的资源,以构建大肠杆菌的凝集素-聚糖相互作用网络。另外,电子膜组管线可以扩展到需要解决宿主病原体相互作用的其他类似生物系统。











































 京公网安备 11010802027423号
京公网安备 11010802027423号