当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Coupling Coarse-Grained to Fine-Grained Models via Hamiltonian Replica Exchange.
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2020-06-22 , DOI: 10.1021/acs.jctc.0c00429 Yang Liu 1 , Weria Pezeshkian 1 , Jonathan Barnoud 1 , Alex H de Vries 1 , Siewert J Marrink 1
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2020-06-22 , DOI: 10.1021/acs.jctc.0c00429 Yang Liu 1 , Weria Pezeshkian 1 , Jonathan Barnoud 1 , Alex H de Vries 1 , Siewert J Marrink 1
Affiliation
|
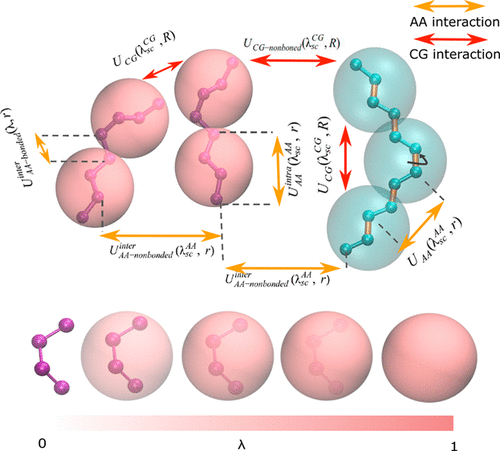
The energy landscape of biomolecular systems contains many local minima that are separated by high energy barriers. Sampling this landscape in molecular dynamics simulations is a challenging task and often requires the use of enhanced sampling techniques. Here, we increase the sampling efficiency by coupling the fine-grained (FG) GROMOS force field to the coarse-grained (CG) Martini force field via the Hamiltonian replica exchange method (HREM). We tested the efficiency of this procedure using a lutein/octane system. In traditional simulations, cis–trans transitions of lutein are barely observed due to the high energy barrier separating these states. However, many of these transitions are sampled with our HREM scheme. The proposed method offers new possibilities for enhanced sampling of biomolecular conformations, making use of CG models without compromising the accuracy of the FG model.
中文翻译:

通过汉密尔顿副本交换将粗粒模型耦合到细粒模型。
生物分子系统的能量格局包含许多局部最小值,这些最小值被高能垒隔开。在分子动力学模拟中对这种情况进行采样是一项艰巨的任务,通常需要使用增强的采样技术。在这里,我们通过汉密尔顿复制副本交换方法(HREM)将细粒(FG)GROMOS力场耦合到粗粒(CG)马蒂尼力场来提高采样效率。我们使用叶黄素/辛烷体系测试了该程序的效率。在传统的模拟中,由于分隔这些状态的高能垒,几乎未观察到叶黄素的顺式-反式转变。但是,其中许多过渡都是通过我们的HREM方案进行采样的。所提出的方法为增强生物分子构象的采样提供了新的可能性,
更新日期:2020-08-11
中文翻译:

通过汉密尔顿副本交换将粗粒模型耦合到细粒模型。
生物分子系统的能量格局包含许多局部最小值,这些最小值被高能垒隔开。在分子动力学模拟中对这种情况进行采样是一项艰巨的任务,通常需要使用增强的采样技术。在这里,我们通过汉密尔顿复制副本交换方法(HREM)将细粒(FG)GROMOS力场耦合到粗粒(CG)马蒂尼力场来提高采样效率。我们使用叶黄素/辛烷体系测试了该程序的效率。在传统的模拟中,由于分隔这些状态的高能垒,几乎未观察到叶黄素的顺式-反式转变。但是,其中许多过渡都是通过我们的HREM方案进行采样的。所提出的方法为增强生物分子构象的采样提供了新的可能性,











































 京公网安备 11010802027423号
京公网安备 11010802027423号