当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Predicting Binding from Screening Assays with Transformer Network Embeddings.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-06-22 , DOI: 10.1021/acs.jcim.9b01212 Paul Morris 1 , Rachel St Clair 1 , William Edward Hahn 1 , Elan Barenholtz 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-06-22 , DOI: 10.1021/acs.jcim.9b01212 Paul Morris 1 , Rachel St Clair 1 , William Edward Hahn 1 , Elan Barenholtz 1
Affiliation
|
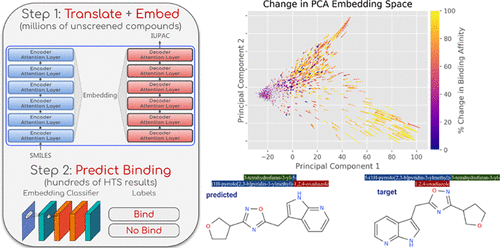
Cheminformatics aims to assist in chemistry applications that depend on molecular interactions, structural characteristics, and functional properties. The arrival of deep learning and the abundance of easily accessible chemical data from repositories like PubChem have enabled advancements in computer-aided drug discovery. Virtual high-throughput screening (vHTS) is one such technique that integrates chemical domain knowledge to perform in silico biomolecular simulations, but prediction of binding affinity is restricted due to limited availability of ground-truth binding assay results. Here, text representations of 83 000 000 molecules are leveraged to perform single-target binding affinity prediction directly on the outcome of screening assays. The embedding of an end-to-end transformer neural network, trained to encode the structural characteristics of a molecule via a text-based translation task, is repurposed through transfer learning to classify binding affinity to single targets with few known binding compounds. We quantify the observed increase in AUC on binding prediction tasks between classifiers trained on the translation embedding versus those using an untrained embedding. Visualization of the embedding space reveals organization of structural and functional properties that aid binding prediction. The pretrained transformer, data, and associated software to extract embeddings are made publicly available at https://github.com/mpcrlab/MolecularTransformerEmbeddings.
中文翻译:

通过具有变压器网络嵌入的筛选分析来预测结合。
化学信息学旨在协助依赖分子相互作用,结构特征和功能特性的化学应用。深度学习的到来以及来自诸如PubChem之类的存储库的大量易于访问的化学数据,已使计算机辅助药物发现得到了发展。虚拟高通量筛选(vHTS)是整合化学领域知识以进行计算机生物分子模拟的一种此类技术,但由于地面真相结合测定结果的可用性有限,因此对结合亲和力的预测受到限制。在此,直接利用筛选测定的结果来利用83000000个分子的文本表示来执行单靶结合亲和力预测。端到端变压器神经网络的嵌入,通过基于文本的翻译任务来对分子的结构特征进行编码的训练专家,通过转移学习将其重新利用,以利用很少的已知结合化合物对与单个靶标的结合亲和力进行分类。我们对在翻译嵌入训练的分类器与使用未经训练的嵌入的分类器之间的绑定预测任务上的AUC量化进行量化。嵌入空间的可视化揭示了有助于绑定预测的结构和功能属性的组织。可以在https://github.com/mpcrlab/MolecularTransformerEmbeddings上公开获得经过预训练的转换器,数据以及用于提取嵌入的相关软件。我们对在翻译嵌入训练的分类器与使用未经训练的嵌入的分类器之间的绑定预测任务上的AUC量化进行量化。嵌入空间的可视化揭示了有助于绑定预测的结构和功能属性的组织。可以在https://github.com/mpcrlab/MolecularTransformerEmbeddings上公开获得经过预训练的转换器,数据以及用于提取嵌入的相关软件。我们对在翻译嵌入训练的分类器与使用未经训练的嵌入的分类器之间的绑定预测任务上的AUC量化进行量化。嵌入空间的可视化揭示了有助于绑定预测的结构和功能属性的组织。可以在https://github.com/mpcrlab/MolecularTransformerEmbeddings上公开获得经过预训练的转换器,数据以及用于提取嵌入的相关软件。
更新日期:2020-06-22
中文翻译:

通过具有变压器网络嵌入的筛选分析来预测结合。
化学信息学旨在协助依赖分子相互作用,结构特征和功能特性的化学应用。深度学习的到来以及来自诸如PubChem之类的存储库的大量易于访问的化学数据,已使计算机辅助药物发现得到了发展。虚拟高通量筛选(vHTS)是整合化学领域知识以进行计算机生物分子模拟的一种此类技术,但由于地面真相结合测定结果的可用性有限,因此对结合亲和力的预测受到限制。在此,直接利用筛选测定的结果来利用83000000个分子的文本表示来执行单靶结合亲和力预测。端到端变压器神经网络的嵌入,通过基于文本的翻译任务来对分子的结构特征进行编码的训练专家,通过转移学习将其重新利用,以利用很少的已知结合化合物对与单个靶标的结合亲和力进行分类。我们对在翻译嵌入训练的分类器与使用未经训练的嵌入的分类器之间的绑定预测任务上的AUC量化进行量化。嵌入空间的可视化揭示了有助于绑定预测的结构和功能属性的组织。可以在https://github.com/mpcrlab/MolecularTransformerEmbeddings上公开获得经过预训练的转换器,数据以及用于提取嵌入的相关软件。我们对在翻译嵌入训练的分类器与使用未经训练的嵌入的分类器之间的绑定预测任务上的AUC量化进行量化。嵌入空间的可视化揭示了有助于绑定预测的结构和功能属性的组织。可以在https://github.com/mpcrlab/MolecularTransformerEmbeddings上公开获得经过预训练的转换器,数据以及用于提取嵌入的相关软件。我们对在翻译嵌入训练的分类器与使用未经训练的嵌入的分类器之间的绑定预测任务上的AUC量化进行量化。嵌入空间的可视化揭示了有助于绑定预测的结构和功能属性的组织。可以在https://github.com/mpcrlab/MolecularTransformerEmbeddings上公开获得经过预训练的转换器,数据以及用于提取嵌入的相关软件。











































 京公网安备 11010802027423号
京公网安备 11010802027423号