Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structural Basis of Nucleosome Recognition and Modulation.
BioEssays ( IF 3.2 ) Pub Date : 2020-06-22 , DOI: 10.1002/bies.201900234 Rajivgandhi Sundaram 1, 2 , Dileep Vasudevan 1
BioEssays ( IF 3.2 ) Pub Date : 2020-06-22 , DOI: 10.1002/bies.201900234 Rajivgandhi Sundaram 1, 2 , Dileep Vasudevan 1
Affiliation
|
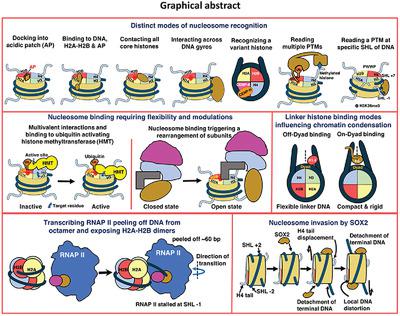
Chromatin structure and dynamics regulate key cellular processes such as DNA replication, transcription, repair, remodeling, and gene expression, wherein different protein factors interact with the nucleosomes. In these events, DNA and RNA polymerases, chromatin remodeling enzymes and transcription factors interact with nucleosomes, either in a DNA‐sequence‐specific manner and/or by recognizing different structural features on the nucleosome. The molecular details of the recognition of a nucleosome by different viral proteins, remodeling enzymes, histone post‐translational modifiers, and RNA polymerase II, have been explored in the recent past. The present review puts forth critical insights into the basic mechanisms of nucleosome recognition by the various protein factors and the role of distinct surface epitopes on a nucleosome. These determinants of the underlying specificity include features such as the acidic patch, arginine anchor, histone post‐translational modifications, core DNA, DNA lesions, and linker DNA.
中文翻译:

核小体识别和调节的结构基础。
染色质结构和动力学调节关键的细胞过程,如 DNA 复制、转录、修复、重塑和基因表达,其中不同的蛋白质因子与核小体相互作用。在这些事件中,DNA 和 RNA 聚合酶、染色质重塑酶和转录因子以 DNA 序列特异性方式和/或通过识别核小体上的不同结构特征与核小体相互作用。最近已经探索了不同病毒蛋白、重塑酶、组蛋白翻译后修饰剂和 RNA 聚合酶 II 识别核小体的分子细节。本综述对各种蛋白质因子识别核小体的基本机制以及不同表面表位在核小体上的作用提出了重要见解。
更新日期:2020-08-26
中文翻译:

核小体识别和调节的结构基础。
染色质结构和动力学调节关键的细胞过程,如 DNA 复制、转录、修复、重塑和基因表达,其中不同的蛋白质因子与核小体相互作用。在这些事件中,DNA 和 RNA 聚合酶、染色质重塑酶和转录因子以 DNA 序列特异性方式和/或通过识别核小体上的不同结构特征与核小体相互作用。最近已经探索了不同病毒蛋白、重塑酶、组蛋白翻译后修饰剂和 RNA 聚合酶 II 识别核小体的分子细节。本综述对各种蛋白质因子识别核小体的基本机制以及不同表面表位在核小体上的作用提出了重要见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号