当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Bayesian Active Learning for Optimization and Uncertainty Quantification in Protein Docking.
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2020-06-19 , DOI: 10.1021/acs.jctc.0c00476 Yue Cao 1 , Yang Shen 1, 2
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2020-06-19 , DOI: 10.1021/acs.jctc.0c00476 Yue Cao 1 , Yang Shen 1, 2
Affiliation
|
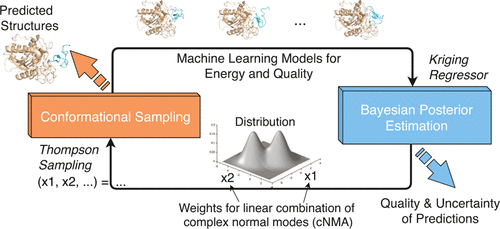
Ab initio protein docking represents a major challenge for optimizing a noisy and costly “black box”-like function in a high-dimensional space. Despite progress in this field, there is a lack of rigorous uncertainty quantification (UQ). To fill the gap, we introduce a novel algorithm, Bayesian active learning (BAL), for optimization and UQ of such black-box functions with applications to flexible protein docking. BAL directly models the posterior distribution of the global optimum (i.e., native structures) with active sampling and posterior estimation iteratively feeding each other. Furthermore, it uses complex normal modes to span a homogeneous, Euclidean conformation space suitable for high-dimensional optimization and constructs funnel-like energy models for quality estimation of encounter complexes. Over a protein-docking benchmark set and a CAPRI set including homology docking, we establish that BAL significantly improves against starting points from rigid docking and refinements by particle swarm optimization, providing a top-3 near-native prediction for one third targets. Quality assessment empowered with UQ leads to tight quality intervals with half range around 25% of the actual interface root-mean-square deviation and confidence level at 85%. BAL’s estimated probability of a prediction being near-native achieves binary classification AUROC at 0.93 and area under the precision recall curve over 0.60 (compared to 0.50 and 0.14, respectively, by chance), which also improves ranking predictions. This study represents the first UQ solution for protein docking, with rigorous theoretical frameworks and comprehensive empirical assessments.
中文翻译:

用于蛋白质对接中优化和不确定性量化的贝叶斯主动学习。
Ab initio蛋白质对接代表了在高维空间中优化嘈杂且昂贵的“黑匣子”式功能的主要挑战。尽管该领域取得了进展,但缺乏严格的不确定性量化 (UQ)。为了填补这一空白,我们引入了一种新颖的算法,即贝叶斯主动学习 (BAL),用于优化和 UQ 这种黑盒函数,并应用于灵活的蛋白质对接。BAL 直接模拟全局最优值的后验分布(即,原生结构)具有主动采样和后验估计迭代地相互馈送。此外,它使用复杂的正常模式来跨越适用于高维优化的均匀欧几里得构象空间,并构建漏斗状能量模型来估计遭遇复合物的质量。在蛋白质对接基准集和包括同源对接的 CAPRI 集上,我们确定 BAL 相对于刚性对接和粒子群优化改进的起点有显着改善,为三分之一的目标提供了前 3 名的近原生预测。使用 UQ 进行的质量评估导致质量区间很窄,其一半范围约为实际界面均方根偏差的 25%,置信水平为 85%。BAL 对接近原生的预测的估计概率实现了 0.93 的二元分类 AUROC 和超过 0.60 的精确召回曲线下面积(与分别为 0.50 和 0.14 相比,偶然),这也提高了排名预测。这项研究代表了 UQ 的第一个蛋白质对接解决方案,具有严格的理论框架和全面的经验评估。
更新日期:2020-08-11
中文翻译:

用于蛋白质对接中优化和不确定性量化的贝叶斯主动学习。
Ab initio蛋白质对接代表了在高维空间中优化嘈杂且昂贵的“黑匣子”式功能的主要挑战。尽管该领域取得了进展,但缺乏严格的不确定性量化 (UQ)。为了填补这一空白,我们引入了一种新颖的算法,即贝叶斯主动学习 (BAL),用于优化和 UQ 这种黑盒函数,并应用于灵活的蛋白质对接。BAL 直接模拟全局最优值的后验分布(即,原生结构)具有主动采样和后验估计迭代地相互馈送。此外,它使用复杂的正常模式来跨越适用于高维优化的均匀欧几里得构象空间,并构建漏斗状能量模型来估计遭遇复合物的质量。在蛋白质对接基准集和包括同源对接的 CAPRI 集上,我们确定 BAL 相对于刚性对接和粒子群优化改进的起点有显着改善,为三分之一的目标提供了前 3 名的近原生预测。使用 UQ 进行的质量评估导致质量区间很窄,其一半范围约为实际界面均方根偏差的 25%,置信水平为 85%。BAL 对接近原生的预测的估计概率实现了 0.93 的二元分类 AUROC 和超过 0.60 的精确召回曲线下面积(与分别为 0.50 和 0.14 相比,偶然),这也提高了排名预测。这项研究代表了 UQ 的第一个蛋白质对接解决方案,具有严格的理论框架和全面的经验评估。











































 京公网安备 11010802027423号
京公网安备 11010802027423号