Journal of Molecular Biology ( IF 4.7 ) Pub Date : 2020-06-19 , DOI: 10.1016/j.jmb.2020.06.014 Jenna N Beyer 1 , Parisa Hosseinzadeh 2 , Ilana Gottfried-Lee 1 , Elise M Van Fossen 1 , Phillip Zhu 1 , Riley M Bednar 1 , P Andrew Karplus 1 , Ryan A Mehl 1 , Richard B Cooley 1
|
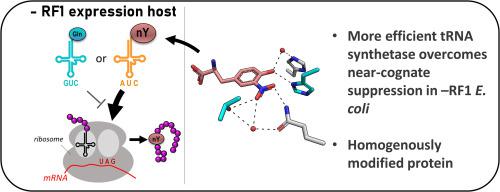
Genetic code expansion (GCE) technologies incorporate non-canonical amino acids (ncAAs) into proteins at amber stop codons. To avoid unwanted truncated protein and improve ncAA–protein yields, genomically recoded strains of Escherichia coli lacking Release Factor 1 (RF1) are becoming increasingly popular expression hosts for GCE applications. In the absence of RF1, however, endogenous near-cognate amber suppressing tRNAs can lead to contaminating protein forms with natural amino acids in place of the ncAA. Here, we show that a second-generation amino-acyl tRNA synthetase (aaRS)/tRNACUA pair for site-specific incorporation of 3-nitro-tyrosine could not outcompete near-cognate suppression in an RF1-deficient expression host and therefore could not produce homogenously nitrated protein. To resolve this, we used Rosetta to target positions in the nitroTyr aaRS active site for improved substrate binding, and then constructed of a small library of variants to subject to standard selection protocols. The top selected variant had an ~ 2-fold greater efficiency, and remarkably, this relatively small improvement enabled homogeneous incorporation of nitroTyr in an RF1-deficient expression host and thus eliminates truncation issues associated with typical RF1-containing expression hosts. Structural and biochemical data suggest the aaRS efficiency improvement is based on higher affinity substrate binding. Taken together, the modest improvement in aaRS efficiency provides a large practical impact and expands our ability to study the role protein nitration plays in disease development through producing homogenous, truncation-free nitroTyr-containing protein. This work establishes Rosetta-guided design and incremental aaRS improvement as a viable and accessible path to improve GCE systems challenged by truncation and/or near-cognate suppression issues.
中文翻译:

用改良的硝基酪氨酸tRNA合成酶克服释放因子1-缺陷型宿主中的近齿抑制。
遗传密码扩展(GCE)技术在琥珀色终止密码子中将非规范氨基酸(ncAAs)整合到蛋白质中。为了避免不想要的截短的蛋白并提高ncAA-蛋白的产量,缺少释放因子1(RF1)的大肠埃希菌的基因组重新编码菌株正成为GCE应用中越来越受欢迎的表达宿主。然而,在没有RF1的情况下,内源性近同源琥珀色抑制性tRNA可能导致天然氨基酸代替ncAA污染蛋白质形式。在这里,我们显示了第二代氨基酰基tRNA合成酶(aaRS)/ tRNA CUA一对位点特异性结合3-硝基酪氨酸在RF1缺乏表达宿主中不能超过近同源抑制,因此不能产生均一的硝化蛋白质。为了解决这个问题,我们使用Rosetta靶向了nitroTyr aaRS活性位点中的位置,以改善底物结合,然后构建了一个小的变异库来接受标准选择方案。排在最前面的变体的效率提高了约2倍,并且值得注意的是,这种相对较小的改进使nitroTyr在RF1缺陷型表达宿主中均匀掺入,从而消除了与典型的含RF1的表达宿主相关的截短问题。结构和生化数据表明,aaRS效率的提高是基于更高的亲和性底物结合。在一起 aaRS效率的适度提高将产生巨大的实际影响,并扩大我们研究蛋白质硝化通过产生均质,无截短的含硝基酪氨酸的蛋白质在疾病发展中所起的作用的能力。这项工作建立了罗塞塔指导的设计和对aaRS的逐步改进,这是一条可行且可访问的途径,可改善受到截断和/或近同源抑制问题挑战的GCE系统。











































 京公网安备 11010802027423号
京公网安备 11010802027423号