Journal of Neuroscience Methods ( IF 2.7 ) Pub Date : 2020-06-18 , DOI: 10.1016/j.jneumeth.2020.108804 Xinglong Wu 1 , Shangbin Chen 2 , Jin Huang 3 , Anan Li 2 , Rong Xiao 2 , Xinwu Cui 4
|
Background
Deep learning models are turning out to be increasingly popular in biomedical image processing. The fruitful utilization of these models, in most cases, is substantially restricted by the complicated configuration of computational environments, resulting in the noteworthy increment of the time and endeavors to reproduce the outcomes of the models.
New Method
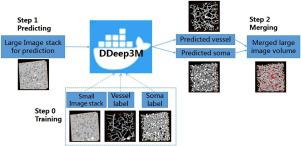
We thus present a Docker-based method for better use of deep learning models and quicker reproduction of model performance for multiple data sources, permitting progressively more biomedical scientists to attempt the new technology conveniently in their domain. Here, we introduce a Docker-powered deep learning model, named as DDeep3M and validated it with the electron microscopy data volumes (microscale).
Results
DDeep3M is utilized to the 3D optical microscopy image stack in mouse brain for the image segmentation (mesoscale). It achieves high accuracy on both vessels and somata structures with all the recall/precision scores and Dice indexes over 0.96. DDeep3M also reports the state-of-the-art performance in the MRI data (macroscale) for brain tumor segmentation.
Comparison with existing methods
We compare the performance and efficiency of DDeep3M with three existing models on image datasets varying from micro- to macro-scales.
Conclusion
DDeep3M is a friendly, convenient and efficient tool for image segmentations in biomedical research. DDeep3M is open sourced with the codes and pretrained model weights available at https://github.com/cakuba/DDeep3m.
中文翻译:

DDeep3M:Docker支持的深度学习,用于生物医学图像分割。
背景
深度学习模型在生物医学图像处理中越来越受欢迎。在大多数情况下,对这些模型的卓有成效的利用在很大程度上受到计算环境复杂配置的限制,从而导致时间的显着增加以及努力重现模型的结果。
新方法
因此,我们提出了一种基于Docker的方法,可以更好地利用深度学习模型并更快地重现多个数据源的模型性能,从而使越来越多的生物医学科学家能够方便地在其领域尝试新技术。在这里,我们介绍了一个基于Docker的深度学习模型,该模型名为DDeep3M,并已通过电子显微镜数据量(微尺度)进行了验证。
结果
DDeep3M用于鼠标大脑中的3D光学显微镜图像堆栈,以进行图像分割(中尺度)。它的所有召回/精确度得分和Dice指数均超过0.96,在血管和躯体结构上均达到了高精度。DDeep3M还在MRI数据(宏量表)中报告了脑肿瘤分割的最新技术。
与现有方法的比较
我们将DDeep3M的性能和效率与从微观到宏观的图像数据集上的三个现有模型进行了比较。
结论
DDeep3M是用于生物医学研究中图像分割的友好,便捷和高效的工具。DDeep3M是开源的,其代码和预训练的模型权重可从https://github.com/cakuba/DDeep3m获得。










































 京公网安备 11010802027423号
京公网安备 11010802027423号