当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Improved Parameterization of Protein-DNA Interactions for Molecular Dynamics Simulations of PCNA Diffusion on DNA.
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2020-06-16 , DOI: 10.1021/acs.jctc.0c00241 Seonju You 1 , Hong-Guen Lee 2, 3 , Kimoon Kim 1, 3 , Jejoong Yoo 3, 4
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2020-06-16 , DOI: 10.1021/acs.jctc.0c00241 Seonju You 1 , Hong-Guen Lee 2, 3 , Kimoon Kim 1, 3 , Jejoong Yoo 3, 4
Affiliation
|
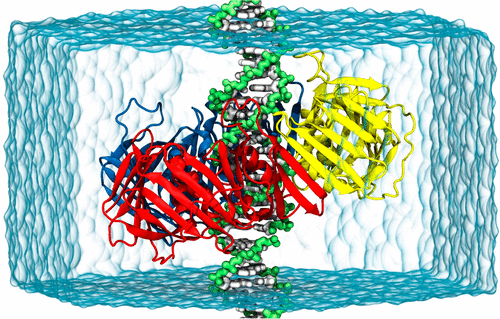
As the field of molecular dynamics simulation utilizing the force fields is moving toward more complex systems, the accuracy of intermolecular interactions has become a central issue of the field. Here, we quantitatively evaluate the accuracy of the protein–DNA interactions in AMBER and CHARMM force fields by comparing experimental and simulated diffusion coefficients of proliferating cell nuclear antigen. We find that both force fields underestimate diffusion coefficients by at least an order of magnitude because the interactions between basic amino acids and DNA phosphate groups are too attractive. Then, we propose Lennard-Jones parameters optimized using the experimental osmotic pressure data of model chemicals, by using which one can reproduce the experimental diffusion coefficients. Newly optimized parameters will have a broad impact on general protein–DNA interactions.
中文翻译:

蛋白质-DNA相互作用的改进参数化,用于PCNA在DNA上扩散的分子动力学模拟。
随着利用力场的分子动力学模拟领域向更复杂的系统发展,分子间相互作用的准确性已成为该领域的中心问题。在这里,我们通过比较增殖细胞核抗原的实验扩散系数和模拟扩散系数,定量评估AMBER和CHARMM力场中蛋白质与DNA相互作用的准确性。我们发现两个力场都低估了扩散系数至少一个数量级,因为碱性氨基酸和DNA磷酸基团之间的相互作用太吸引人了。然后,我们提出了使用模型化学品的实验渗透压数据优化的Lennard-Jones参数,从而可以再现实验扩散系数。
更新日期:2020-07-14
中文翻译:

蛋白质-DNA相互作用的改进参数化,用于PCNA在DNA上扩散的分子动力学模拟。
随着利用力场的分子动力学模拟领域向更复杂的系统发展,分子间相互作用的准确性已成为该领域的中心问题。在这里,我们通过比较增殖细胞核抗原的实验扩散系数和模拟扩散系数,定量评估AMBER和CHARMM力场中蛋白质与DNA相互作用的准确性。我们发现两个力场都低估了扩散系数至少一个数量级,因为碱性氨基酸和DNA磷酸基团之间的相互作用太吸引人了。然后,我们提出了使用模型化学品的实验渗透压数据优化的Lennard-Jones参数,从而可以再现实验扩散系数。



























 京公网安备 11010802027423号
京公网安备 11010802027423号