Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-05-01 , DOI: 10.2174/1574893614666191127101836 Juan Liu 1 , Xinjie Lian 1 , Feng Liu 1 , Xueling Yan 1 , Chunyan Cheng 1 , Lijia Cheng 1 , Xiaolin Sun 2 , Zheng Shi 1
|
Background: Oral Squamous Cell Carcinoma (OSCC) is the most common malignant epithelial neoplasm. It is located within the top 10 ranking incidence of cancers with a poor prognosis and low survival rates. New breakthroughs of therapeutic strategies are therefore needed to improve the survival rate of OSCC harboring patients.
Objective: Since targeted therapy is considered as the most promising therapeutic strategies in cancer, it is of great significance to identify novel targets and drugs for the treatment of OSCC.
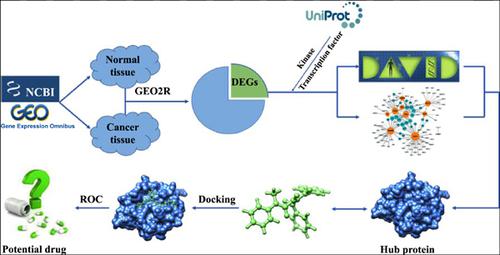
Methods: A series of bioinformatics approaches were launched to identify the hub proteins and their potential agents. Microarray analysis and several online functional activity network analysis were firstly utilized to recognize drug targets in OSCC. Subsequently, molecular docking was used to screen their potential drugs from the specs chemistry database. At the same time, the assessment of ligand-based virtual screening model was also evaluated.
Results: In this study, two microarray data (GSE31056, GSE23558) were firstly selected and analyzed to get consensus candidate genes including 681 candidate genes. Additionally, we selected 33 candidate genes based on whether they belong to the kinases and transcription factors and further clustered candidate hub targets based on functions and signaling pathways with significant enrichment analysis by using DAVID and STRING online databases. Then, core PPI network was then identified and we manually selected GRB2 and IGF1 as the key drug targets according to the network analysis and previous references. Lastly, virtual screening was performed to identify potential small molecules which could target these two targets, and such small molecules can serve as the promising candidate agents for future drug development.
Conclusion: In summary, our study might provide novel insights for understanding of the underlying molecular events of OSCC, and our discovered candidate targets and candidate agents could be used as the promising therapeutic strategies for the treatment of OSCC.
中文翻译:

口腔鳞状细胞癌中新型关键靶标和候选药物的鉴定
背景:口腔鳞状细胞癌(OSCC)是最常见的恶性上皮肿瘤。它位于预后较差,生存率较低的癌症排名前10位。因此,需要新的治疗策略突破,以提高OSCC病人的生存率。
目的:由于靶向治疗被认为是癌症中最有前途的治疗策略,因此确定新的靶向治疗OSCC的靶标和药物具有重要意义。
方法:推出了一系列生物信息学方法来鉴定轮毂蛋白及其潜在作用因子。首先利用微阵列分析和一些在线功能活动网络分析来识别OSCC中的药物靶标。随后,分子对接用于从规格化学数据库中筛选其潜在药物。同时,还评估了基于配体的虚拟筛选模型的评估。
结果:在这项研究中,首先选择并分析了两个微阵列数据(GSE31056,GSE23558)以得到共有候选基因,包括681个候选基因。此外,我们根据它们是否属于激酶和转录因子选择了33个候选基因,并使用DAVID和STRING在线数据库根据功能和信号途径进一步聚类了候选枢纽靶标,并进行了显着的富集分析。然后,确定核心PPI网络,并根据网络分析和先前的参考文献,手动选择GRB2和IGF1作为关键药物靶标。最后,进行了虚拟筛选以识别可能靶向这两个靶标的潜在小分子,并且这些小分子可以用作未来药物开发的有希望的候选药物。
结论:总的来说,我们的研究可能为了解OSCC的潜在分子事件提供新的见解,并且我们发现的候选靶标和候选药物可以用作OSCC的有前途的治疗策略。










































 京公网安备 11010802027423号
京公网安备 11010802027423号