Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-05-01 , DOI: 10.2174/1574893614666191105160633 Abdelbasset Boukelia 1 , Anouar Boucheham 2 , Meriem Belguidoum 1 , Mohamed Batouche 3 , Farida Zehraoui 4 , Fariza Tahi 4
|
Background: Molecular biomarkers show new ways to understand many disease processes. Noncoding RNAs as biomarkers play a crucial role in several cellular activities, which are highly correlated to many human diseases especially cancer. The classification and the identification of ncRNAs have become a critical issue due to their application, such as biomarkers in many human diseases.
Objective: Most existing computational tools for ncRNA classification are mainly used for classifying only one type of ncRNA. They are based on structural information or specific known features. Furthermore, these tools suffer from a lack of significant and validated features. Therefore, the performance of these methods is not always satisfactory.
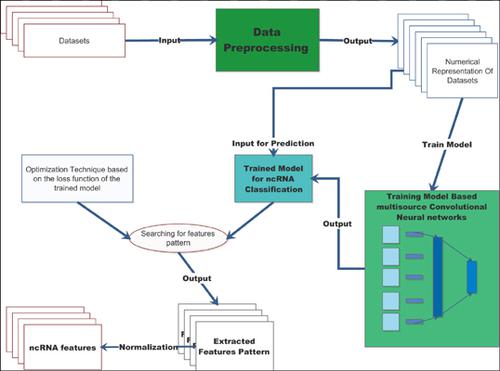
Methods: We propose a novel approach named imCnC for ncRNA classification based on multisource deep learning, which integrates several data sources such as genomic and epigenomic data to identify several ncRNA types. Also, we propose an optimization technique to visualize the extracted features pattern from the multisource CNN model to measure the epigenomics features of each ncRNA type.
Results: The computational results using a dataset of 16 human ncRNA classes downloaded from RFAM show that imCnC outperforms the existing tools. Indeed, imCnC achieved an accuracy of 94,18%. In addition, our method enables to discover new ncRNA features using an optimization technique to measure and visualize the features pattern of the imCnC classifier.
中文翻译:

基于深度学习的非编码RNA分类的一种集成方法
背景:分子生物标记物显示了理解许多疾病过程的新方法。作为生物标志物的非编码RNA在多种细胞活动中起着至关重要的作用,这些活动与许多人类疾病尤其是癌症高度相关。由于ncRNA的应用,例如许多人类疾病中的生物标志物,它们的分类和鉴定已成为一个关键问题。
目的:目前大多数用于ncRNA分类的计算工具主要用于仅对一种类型的ncRNA进行分类。它们基于结构信息或特定的已知功能。此外,这些工具缺少重要且经过验证的功能。因此,这些方法的性能并不总是令人满意。
方法:我们提出了一种基于多源深度学习的名为imCnC的ncRNA分类新方法,该方法整合了诸如基因组和表观基因组数据之类的多种数据源,以识别多种ncRNA类型。此外,我们提出了一种优化技术,以可视化方式从多源CNN模型中提取特征模式,以测量每种ncRNA类型的表观基因组学特征。
结果:使用从RFAM下载的16种人类ncRNA类的数据集得出的计算结果表明,imCnC的性能优于现有工具。实际上,imCnC的准确度达到94.18%。另外,我们的方法能够使用优化技术来发现新的ncRNA特征,以测量和可视化imCnC分类器的特征模式。











































 京公网安备 11010802027423号
京公网安备 11010802027423号