Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-05-01 , DOI: 10.2174/1574893614666191105155713 Huaixu Zhu 1 , Xiuquan Du 1 , Yu Yao 1
|
Background/Objective: Protein-protein interactions are essentials for most cellular processes and thus, unveiling how proteins interact with is a crucial question that can be better understood by recognizing which residues participate in the interaction. Although many computational approaches have been proposed to predict interface residues, their feature perspective and model learning ability are not enough to achieve ideal results. So, our objective is to improve the predictive performance under considering feature perspective and new learning algorithm.
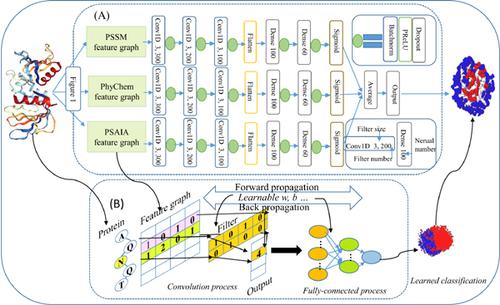
Method: In this study, we proposed an ensemble deep convolutional neural network, which explores the context and positional context of consecutive residues within a protein sub-sequence. Specifically, unlike the feature view of previous methods, ConvsPPIS uses evolutionary, physicochemical, and structural protein characteristics to construct their own feature graph respectively. After that, three independent deep convolutional neural networks are trained on each type of feature graph for learning the underlying pattern in sub-sequence. Lastly, we integrated those three deep networks into an ensemble predictor with leveraging complementary information of those features to predict potential interface residues.
Results: Some comparative experiments have conducted through 10-fold cross-validation. The results indicated that ConvsPPIS achieved superior performance on DBv5-Sel dataset with an accuracy of 88%. Additional experiments on CAPRI-Alone dataset demonstrated ConvsPPIS has also better prediction performance.
Conclusion: The ConvsPPIS method provided a new perspective to capture protein feature expression for identifying protein-protein interaction sites. The results proved the superiority of this method.
中文翻译:

ConvsPPIS:通过具有特征图的集成卷积神经网络识别蛋白质-蛋白质相互作用位点
背景/目的:蛋白质-蛋白质相互作用是大多数细胞过程必不可少的,因此,揭示蛋白质如何相互作用是一个至关重要的问题,可以通过识别哪些残基参与相互作用来更好地理解。尽管已经提出了许多计算方法来预测界面残差,但是它们的特征角度和模型学习能力不足以实现理想的结果。因此,我们的目标是在考虑特征角度和新的学习算法的情况下提高预测性能。
方法:在这项研究中,我们提出了一个集成的深度卷积神经网络,该网络探索蛋白质子序列中连续残基的上下文和位置上下文。具体而言,与以前方法的特征视图不同,ConvsPPIS利用进化,物理化学和结构蛋白特征分别构建自己的特征图。之后,针对每种类型的特征图训练三个独立的深度卷积神经网络,以学习子序列中的基础模式。最后,我们利用这三个特征的互补信息将这三个深度网络集成到一个整体预测器中,以预测潜在的界面残基。
结果:一些比较实验通过10倍交叉验证进行。结果表明,ConvsPPIS在DBv5-Sel数据集上具有卓越的性能,准确性为88%。在CAPRI-Alone数据集上进行的其他实验表明,ConvsPPIS还具有更好的预测性能。
结论:ConvsPPIS方法为捕获蛋白质特征表达以鉴定蛋白质-蛋白质相互作用位点提供了新的视角。结果证明了该方法的优越性。









































 京公网安备 11010802027423号
京公网安备 11010802027423号