当前位置:
X-MOL 学术
›
ACS Comb. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Denoising DNA Encoded Library Screens with Sparse Learning.
ACS Combinatorial Science Pub Date : 2020-06-12 , DOI: 10.1021/acscombsci.0c00007 Péter Kómár 1 , Marko Kalinić 2
ACS Combinatorial Science Pub Date : 2020-06-12 , DOI: 10.1021/acscombsci.0c00007 Péter Kómár 1 , Marko Kalinić 2
Affiliation
|
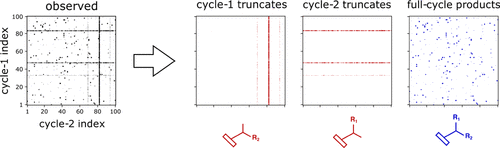
DNA-encoded libraries (DELs) are large, pooled collections of compounds in which every library member is attached to a stretch of DNA encoding its complete synthetic history. DEL-based hit discovery involves affinity selection of the library against a protein of interest, whereby compounds retained by the target are subsequently identified by next-generation sequencing of the corresponding DNA tags. When analyzing the resulting data, one typically assumes that sequencing output (i.e., read counts) is proportional to the binding affinity of a given compound, thus enabling hit prioritization and elucidation of any underlying structure–activity relationships (SAR). This assumption, though, tends to be severely confounded by a number of factors, including variable reaction yields, presence of incomplete products masquerading as their intended counterparts, and sequencing noise. In practice, these confounders are often ignored, potentially contributing to low hit validation rates, and universally leading to loss of valuable information. To address this issue, we have developed a method for comprehensively denoising DEL selection outputs. Our method, dubbed “deldenoiser”, is based on sparse learning and leverages inputs that are commonly available within a DEL generation and screening workflow. Using simulated and publicly available DEL affinity selection data, we show that “deldenoiser” is not only able to recover and rank true binders much more robustly than read count-based approaches but also that it yields scores, which accurately capture the underlying SAR. The proposed method can, thus, be of significant utility in hit prioritization following DEL screens.
中文翻译:

通过稀疏学习对DNA编码的库屏幕进行消噪。
DNA编码文库(DEL)是化合物的大型合并集合,其中每个文库成员均与一段编码其完整合成历史的DNA相连。基于DEL的命中发现涉及对目标蛋白质的亲和力选择,从而随后通过相应DNA标签的下一代测序来鉴定靶标保留的化合物。在分析结果数据时,通常会假设测序输出(即读取计数)与给定化合物的结合亲和力成正比,因此可以按优先顺序排列和阐明任何潜在的结构-活性关系(SAR)。不过,这个假设往往会受到许多因素的严重混淆,包括可变的反应收率,伪装成预期目标的不完整产品的存在,以及排序噪音。在实践中,这些混杂因素通常被忽略,可能导致命中验证率低,并普遍导致有价值信息的丢失。为了解决这个问题,我们开发了一种对DEL选择输出进行全面去噪的方法。我们的方法被称为“ deldenoiser”,是基于稀疏学习并利用DEL生成和筛选工作流程中通常可用的输入。使用模拟的和公开可用的DEL亲和力选择数据,我们表明“ deldenoiser”不仅比基于读取计数的方法能够更可靠地恢复和排列真实的结合物,而且还可以产生得分,从而准确地捕获潜在的SAR。因此,提出的方法可以
更新日期:2020-08-10
中文翻译:

通过稀疏学习对DNA编码的库屏幕进行消噪。
DNA编码文库(DEL)是化合物的大型合并集合,其中每个文库成员均与一段编码其完整合成历史的DNA相连。基于DEL的命中发现涉及对目标蛋白质的亲和力选择,从而随后通过相应DNA标签的下一代测序来鉴定靶标保留的化合物。在分析结果数据时,通常会假设测序输出(即读取计数)与给定化合物的结合亲和力成正比,因此可以按优先顺序排列和阐明任何潜在的结构-活性关系(SAR)。不过,这个假设往往会受到许多因素的严重混淆,包括可变的反应收率,伪装成预期目标的不完整产品的存在,以及排序噪音。在实践中,这些混杂因素通常被忽略,可能导致命中验证率低,并普遍导致有价值信息的丢失。为了解决这个问题,我们开发了一种对DEL选择输出进行全面去噪的方法。我们的方法被称为“ deldenoiser”,是基于稀疏学习并利用DEL生成和筛选工作流程中通常可用的输入。使用模拟的和公开可用的DEL亲和力选择数据,我们表明“ deldenoiser”不仅比基于读取计数的方法能够更可靠地恢复和排列真实的结合物,而且还可以产生得分,从而准确地捕获潜在的SAR。因此,提出的方法可以











































 京公网安备 11010802027423号
京公网安备 11010802027423号