当前位置:
X-MOL 学术
›
Macromolecules
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Force Response of Polypeptide Chains from Water-Explicit MD Simulations
Macromolecules ( IF 5.1 ) Pub Date : 2020-06-12 , DOI: 10.1021/acs.macromol.0c00138 Richard Schwarzl 1 , Susanne Liese 1, 2 , Florian N. Brünig 1 , Fabio Laudisio 1 , Roland R. Netz 1
Macromolecules ( IF 5.1 ) Pub Date : 2020-06-12 , DOI: 10.1021/acs.macromol.0c00138 Richard Schwarzl 1 , Susanne Liese 1, 2 , Florian N. Brünig 1 , Fabio Laudisio 1 , Roland R. Netz 1
Affiliation
|
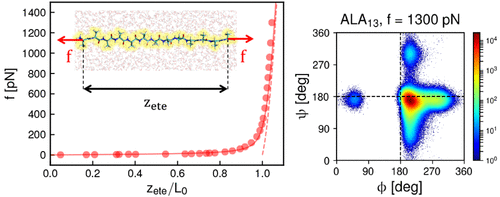
Using molecular dynamics simulations in explicit water, the force–extension relations for the five homopeptides polyglycine, polyalanine, polyasparagine, poly(glutamic acid), and polylysine are investigated. From simulations in the low-force regime the Kuhn length is determined, from simulations in the high-force regime the equilibrium contour length and the linear and nonlinear stretching moduli, which agree well with quantum-chemical density-functional theory calculations, are determined. All these parameters vary considerably between the different polypeptides. The augmented inhomogeneous partially freely rotating chain (iPFRC) model, which accounts for side-chain interactions and restricted dihedral rotation, is demonstrated to describe the simulated force–extension relations very well. We present a quantitative comparison between published experimental single-molecule force–extension curves for different polypeptides with simulation and model predictions. The thermodynamic stretching properties of polypeptides are investigated by decomposition of the stretching free energy into energetic and entropic contributions.
中文翻译:

水显性MD模拟中多肽链的力响应
使用显性水中的分子动力学模拟,研究了五个同肽聚甘氨酸,聚丙氨酸,聚天冬酰胺,聚谷氨酸和聚赖氨酸的力-延伸关系。从低力状态下的模拟确定库恩长度,从高力状态下的模拟确定平衡轮廓长度以及线性和非线性拉伸模量,这与量子化学密度函数理论计算非常吻合。所有这些参数在不同的多肽之间差异很大。增强的非均质部分自由旋转链(iPFRC)模型能够很好地描述模拟的力-延伸关系,该模型考虑了侧链相互作用和受限制的二面角旋转。我们用模拟和模型预测对不同多肽的已发表实验单分子力-延伸曲线进行定量比较。多肽的热力学拉伸特性是通过将拉伸自由能分解为高能和熵的贡献来研究的。
更新日期:2020-06-23
中文翻译:

水显性MD模拟中多肽链的力响应
使用显性水中的分子动力学模拟,研究了五个同肽聚甘氨酸,聚丙氨酸,聚天冬酰胺,聚谷氨酸和聚赖氨酸的力-延伸关系。从低力状态下的模拟确定库恩长度,从高力状态下的模拟确定平衡轮廓长度以及线性和非线性拉伸模量,这与量子化学密度函数理论计算非常吻合。所有这些参数在不同的多肽之间差异很大。增强的非均质部分自由旋转链(iPFRC)模型能够很好地描述模拟的力-延伸关系,该模型考虑了侧链相互作用和受限制的二面角旋转。我们用模拟和模型预测对不同多肽的已发表实验单分子力-延伸曲线进行定量比较。多肽的热力学拉伸特性是通过将拉伸自由能分解为高能和熵的贡献来研究的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号