Structure ( IF 4.4 ) Pub Date : 2020-06-11 , DOI: 10.1016/j.str.2020.05.012 Matthew Sinnott 1 , Sony Malhotra 2 , Mallur Srivatsan Madhusudhan 3 , Konstantinos Thalassinos 1 , Maya Topf 2
|
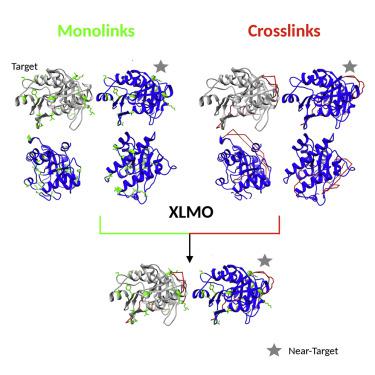
Monolinks are produced in a chemical crosslinking mass spectrometry experiment and are more abundant than crosslinks. They convey residue exposure information, but so far have not been used in the modeling of protein structures. Here, we present the Monolink Depth Score (MoDS), for assessing structural models based on the depth of monolinked residues, corresponding to their distance to the nearest bulk water. Using simulated and reprocessed experimental data from the Proteomic Identification Database, we compare the performance of MoDS to MNXL, our previously developed score for assessing models based on crosslinking data. Our results show that MoDS can be used to effectively score models based on monolinks, and that a crosslink/monolink combined score (XLMO) leads to overall higher performance. The work strongly supports the use of monolink data in the context of integrative structure determination. We also present XLM-Tools, a program to assist in this effort, available at: https://github.com/Topf-Lab/XLM-Tools.
中文翻译:

在蛋白质结构建模中结合来自交联和单联的信息。
单链是在化学交联质谱实验中产生的,比交联更丰富。它们传达残留暴露信息,但迄今为止尚未用于蛋白质结构的建模。在这里,我们提出了 Monolink 深度评分 (MoDS),用于评估基于单链残基深度的结构模型,对应于它们与最近的散装水的距离。使用来自蛋白质组识别数据库的模拟和重新处理的实验数据,我们将 MoDS 的性能与 MNXL 的性能进行比较,MNXL 是我们之前开发的用于评估基于交联数据的模型的分数。我们的结果表明,MoDS 可用于对基于单链路的模型进行有效评分,并且交联/单链路组合评分 (XLMO) 可带来整体更高的性能。这项工作强烈支持在综合结构确定的背景下使用单链数据。我们还介绍了 XLM-Tools,这是一个帮助完成这项工作的程序,可从以下网址获得:https://github.com/Topf-Lab/XLM-Tools。











































 京公网安备 11010802027423号
京公网安备 11010802027423号