Molecular Cell ( IF 14.5 ) Pub Date : 2020-06-11 , DOI: 10.1016/j.molcel.2020.05.025 Pakavarin Louphrasitthiphol 1 , Robert Siddaway 2 , Alessia Loffreda 3 , Vivian Pogenberg 4 , Hans Friedrichsen 2 , Alexander Schepsky 5 , Zhiqiang Zeng 6 , Min Lu 2 , Thomas Strub 7 , Rasmus Freter 2 , Richard Lisle 2 , Eda Suer 2 , Benjamin Thomas 8 , Benjamin Schuster-Böckler 9 , Panagis Filippakopoulos 10 , Mark Middleton 11 , Xin Lu 2 , E Elizabeth Patton 6 , Irwin Davidson 7 , Jean-Philippe Lambert 12 , Matthias Wilmanns 4 , Eiríkur Steingrímsson 13 , Davide Mazza 3 , Colin R Goding 2
|
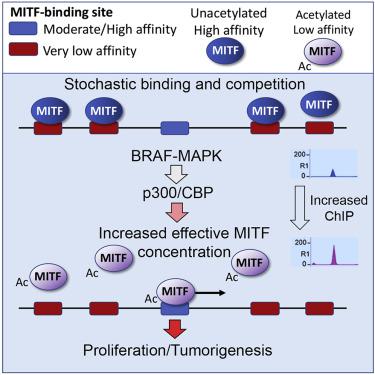
It is widely assumed that decreasing transcription factor DNA-binding affinity reduces transcription initiation by diminishing occupancy of sequence-specific regulatory elements. However, in vivo transcription factors find their binding sites while confronted with a large excess of low-affinity degenerate motifs. Here, using the melanoma lineage survival oncogene MITF as a model, we show that low-affinity binding sites act as a competitive reservoir in vivo from which transcription factors are released by mitogen-activated protein kinase (MAPK)-stimulated acetylation to promote increased occupancy of their regulatory elements. Consequently, a low-DNA-binding-affinity acetylation-mimetic MITF mutation supports melanocyte development and drives tumorigenesis, whereas a high-affinity non-acetylatable mutant does not. The results reveal a paradoxical acetylation-mediated molecular clutch that tunes transcription factor availability via genome-wide redistribution and couples BRAF to tumorigenesis. Our results further suggest that p300/CREB-binding protein-mediated transcription factor acetylation may represent a common mechanism to control transcription factor availability.
中文翻译:

通过乙酰化介导的基因组再分布调整转录因子的可用性。
人们普遍认为,降低转录因子 DNA 结合亲和力会通过减少序列特异性调节元件的占有率来减少转录起始。然而,体内转录因子在遇到大量过量的低亲和力简并基序时会发现它们的结合位点。在这里,使用黑色素瘤谱系存活癌基因 MITF 作为模型,我们表明低亲和力结合位点在体内充当竞争性储存库转录因子通过丝裂原活化蛋白激酶 (MAPK) 刺激的乙酰化释放,以促进其调节元件的占有率增加。因此,低 DNA 结合亲和力乙酰化模拟 MITF 突变支持黑素细胞发育并驱动肿瘤发生,而高亲和力非乙酰化突变体则不支持。结果揭示了一个矛盾的乙酰化介导的分子离合器,它通过全基因组再分配来调节转录因子的可用性,并将 BRAF 与肿瘤发生结合起来。我们的结果进一步表明 p300/CREB 结合蛋白介导的转录因子乙酰化可能代表一种控制转录因子可用性的常见机制。











































 京公网安备 11010802027423号
京公网安备 11010802027423号