Current Biology ( IF 8.1 ) Pub Date : 2020-06-11 , DOI: 10.1016/j.cub.2020.04.070 Kirsten M Ellegaard 1 , Shota Suenami 2 , Ryo Miyazaki 3 , Philipp Engel 1
|
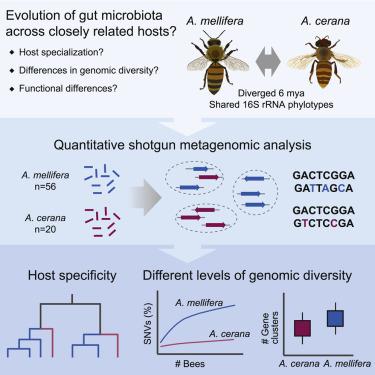
Most bacterial species encompass strains with vastly different gene content. Strain diversity in microbial communities is therefore considered to be of functional importance. Yet little is known about the extent to which related microbial communities differ in diversity at this level and which underlying mechanisms may constrain and maintain strain-level diversity. Here, we used shotgun metagenomics to characterize and compare the gut microbiota of two honey bee species, Apis mellifera and Apis cerana, which diverged about 6 mya. Although the host species are colonized largely by the same bacterial 16S rRNA phylotypes, we find that their communities are host specific when analyzed with genomic resolution. Moreover, despite their similar ecology, A. mellifera displayed a much higher diversity of strains and functional gene content in the microbiota compared to A. cerana, both per colony and per individual bee. In particular, the gene repertoire for polysaccharide degradation was massively expanded in the microbiota of A. mellifera relative to A. cerana. Bee management practices, divergent ecological adaptation, or habitat size may have contributed to the observed differences in microbiota genomic diversity of these key pollinator species. Our results illustrate that the gut microbiota of closely related animal hosts can differ vastly in genomic diversity while displaying similar levels of diversity based on the 16S rRNA gene. Such differences are likely to have consequences for gut microbiota functioning and host-symbiont interactions, highlighting the need for metagenomic studies to understand the ecology and evolution of microbial communities.
中文翻译:

两种密切相关的蜜蜂物种肠道微生物群中菌株水平多样性的巨大差异。
大多数细菌物种包括基因含量差异很大的菌株。因此,微生物群落中的菌株多样性被认为具有功能重要性。然而,人们对相关微生物群落在该水平上的多样性差异程度以及哪些潜在机制可能会限制和维持菌株水平多样性知之甚少。在这里,我们使用鸟枪宏基因组学来表征和比较两种蜜蜂物种(Apis mellifera和Apis cerana)的肠道微生物群,它们的差异大约为 6 米。尽管宿主物种在很大程度上被相同的细菌 16S rRNA 系统型定殖,但我们发现当使用基因组分辨率进行分析时,它们的群落是宿主特异性的。此外,尽管它们的生态相似,A. mellifera与A. cerana相比,每个菌落和每个蜜蜂的微生物群中的菌株和功能基因含量显示出更高的多样性。特别是,相对于A. cerana,A. mellifera的微生物群中多糖降解的基因库大量扩展. 蜜蜂管理实践、不同的生态适应或栖息地大小可能导致观察到的这些关键授粉物种微生物群基因组多样性的差异。我们的结果表明,密切相关的动物宿主的肠道微生物群在基因组多样性方面可能存在很大差异,同时基于 16S rRNA 基因显示出相似的多样性水平。这种差异可能会对肠道微生物群的功能和宿主-共生体相互作用产生影响,突出了宏基因组研究以了解微生物群落的生态和进化的必要性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号