Comparative Biochemistry and Physiology D: Genomics & Proteomics ( IF 2.2 ) Pub Date : 2020-06-10 , DOI: 10.1016/j.cbd.2020.100704 Crisalejandra Rivera-Pérez 1 , Fausto Valenzuela-Quiñonez 1 , Javier Caraveo-Patiño 2
|
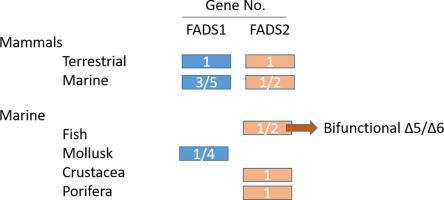
Fatty acid desaturases are key enzymes involved in unsaturated fatty acid biosynthesis, which insert double bonds at specific positions of fatty acids, playing a pivotal role in unsaturated fatty acid synthesis required for membrane lipid fluidity. The ∆5 and ∆6 desaturases are responsible for producing long chain-polyunsaturated fatty acids (LC-PUFA) through their precursors α-linolenic acid and linoleic acid in organisms lacking or with very low ability to synthesize LC-PUFA by themselves. Extensive studies of fatty acid desaturases are available in model organisms, such as humans and mouse; however, the diversity of these genes in the marine biodiversity is less known. This study performed an exhaustive analysis to identify the ∆5 and ∆6 desaturases in the available marine genomes in databases, as well as transcriptomes and EST databases, and their coding sequences were compared to the well-characterized ∆5 and ∆6 desaturases from humans. The FADS1 and FADS2 genetic structures are well conserved among all the organisms analyzed. A common amino acid pattern was identified to discriminate between ∆5 and ∆6 desaturases. The analysis of the conserved motif involved in catalysis showed that 20% of the desaturases, ∆5 and ∆6, have lost motifs required for catalysis. Additionally, bifunctional ∆5/∆6 desaturases were able to be identified by amino acid sequence patterns found in previously described enzymes. A revision of the expression profiles and functional activity on sequences in databases and scientific literature provided information regarding the function of these marine organism enzymes.
中文翻译:

海洋生物脱饱和酶FADS1(∆5)和FADS2(∆6)直向同源物的比较和功能分析。
脂肪酸去饱和酶是参与不饱和脂肪酸生物合成的关键酶,其在脂肪酸的特定位置插入双键,在膜脂质流动性所需的不饱和脂肪酸合成中起关键作用。∆5和∆6去饱和酶负责通过自身的前体α-亚麻酸和亚油酸来生产长链多不饱和脂肪酸(LC-PUFA),而这些生物自身缺乏或合成能力很低。脂肪酸脱氢酶的广泛研究可用于模型生物,例如人和小鼠;但不限于此。然而,这些基因在海洋生物多样性中的多样性却鲜为人知。这项研究进行了详尽的分析,以确定数据库中可用的海洋基因组以及转录组和EST数据库中的Δ5和Δ6去饱和酶,并将它们的编码序列与人类特征明确的∆5和∆6去饱和酶进行比较。在所有分析的生物中,FADS1和FADS2的遗传结构都非常保守。确定了常见的氨基酸模式以区分∆5和∆6去饱和酶。对涉及催化的保守基序的分析表明,有20%的去饱和酶∆5和∆6丢失了催化所需的基序。此外,双功能∆5 / ∆6去饱和酶能够通过前述酶中发现的氨基酸序列模式进行鉴定。数据库和科学文献中序列的表达谱和功能活性的修订提供了有关这些海洋生物酶功能的信息。在所有分析的生物中,FADS1和FADS2的遗传结构都非常保守。确定了常见的氨基酸模式以区分∆5和∆6去饱和酶。对参与催化的保守基序的分析表明,有20%的去饱和酶∆5和∆6丢失了催化所需的基序。此外,还可以通过前述酶中发现的氨基酸序列模式来识别双功能∆5 / ∆6去饱和酶。对数据库和科学文献中序列的表达谱和功能活性的修订提供了有关这些海洋生物酶功能的信息。在所有分析的生物中,FADS1和FADS2的遗传结构都非常保守。确定了常见的氨基酸模式以区分∆5和∆6去饱和酶。对涉及催化的保守基序的分析表明,有20%的去饱和酶∆5和∆6丢失了催化所需的基序。此外,双功能∆5 / ∆6去饱和酶能够通过前述酶中发现的氨基酸序列模式进行鉴定。数据库和科学文献中序列的表达谱和功能活性的修订提供了有关这些海洋生物酶功能的信息。对参与催化的保守基序的分析表明,有20%的去饱和酶∆5和∆6丢失了催化所需的基序。此外,双功能∆5 / ∆6去饱和酶能够通过前述酶中发现的氨基酸序列模式进行鉴定。对数据库和科学文献中序列的表达谱和功能活性的修订提供了有关这些海洋生物酶功能的信息。对参与催化的保守基序的分析表明,有20%的去饱和酶∆5和∆6丢失了催化所需的基序。此外,双功能∆5 / ∆6去饱和酶能够通过前述酶中发现的氨基酸序列模式进行鉴定。对数据库和科学文献中序列的表达谱和功能活性的修订提供了有关这些海洋生物酶功能的信息。











































 京公网安备 11010802027423号
京公网安备 11010802027423号