当前位置:
X-MOL 学术
›
Mol. Omics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
MetaboKit: a comprehensive data extraction tool for untargeted metabolomics.
Molecular Omics ( IF 3.0 ) Pub Date : 2020-06-10 , DOI: 10.1039/d0mo00030b Pradeep Narayanaswamy 1 , Guoshou Teo 2 , Jin Rong Ow 3 , Adam Lau 4 , Philipp Kaldis 5 , Stephen Tate 4 , Hyungwon Choi 6
Molecular Omics ( IF 3.0 ) Pub Date : 2020-06-10 , DOI: 10.1039/d0mo00030b Pradeep Narayanaswamy 1 , Guoshou Teo 2 , Jin Rong Ow 3 , Adam Lau 4 , Philipp Kaldis 5 , Stephen Tate 4 , Hyungwon Choi 6
Affiliation
|
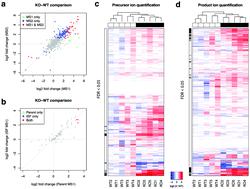
We have developed MetaboKit, a comprehensive software package for compound identification and relative quantification in mass spectrometry-based untargeted metabolomics analysis. In data dependent acquisition (DDA) analysis, MetaboKit constructs a customized spectral library with compound identities from reference spectral libraries, adducts, dimers, in-source fragments (ISF), MS/MS fragmentation spectra, and more importantly the retention time information unique to the chromatography system used in the experiment. Using the customized library, the software performs targeted peak integration for precursor ions in DDA analysis and for precursor and product ions in data independent acquisition (DIA) analysis. With its stringent identification algorithm requiring matches by both MS and MS/MS data, MetaboKit provides identification results with significantly greater specificity than the competing software packages without loss in sensitivity. The proposed MS/MS-based screening of ISFs also reduces the chance of unverifiable identification of ISFs considerably. MetaboKit's quantification module produced peak area values highly correlated with known concentrations in a DIA analysis of the metabolite standards at both MS1 and MS2 levels. Moreover, the analysis of Cdk1Liv−/− mouse livers showed that MetaboKit can identify a wide range of lipid species and their ISFs, and quantitatively reconstitute the well-characterized fatty liver phenotype in these mice. In DIA data, the MS1-level and MS2-level peak area data produced similar fold change estimates in the differential abundance analysis, and the MS2-level peak area data allowed for quantitative comparisons in compounds whose precursor ion chromatogram was too noisy for peak integration.
中文翻译:

MetaboKit:用于非目标代谢组学的全面数据提取工具。
我们已经开发了MetaboKit,这是用于基于质谱的非目标代谢组学分析中化合物鉴定和相对定量的综合软件包。在依赖数据的数据采集(DDA)分析中,MetaboKit使用参考光谱库,加合物,二聚体,源内碎片(ISF),MS / MS碎片光谱,以及更重要的是保留时间信息唯一的化合物身份,构建具有化合物身份的定制光谱库实验中使用的色谱系统。使用定制的库,该软件针对DDA分析中的前体离子以及数据独立获取(DIA)分析中的前体离子和产物离子执行目标峰积分。由于其严格的识别算法要求同时匹配MS和MS / MS数据,与竞争性软件包相比,MetaboKit提供的鉴定结果具有更高的特异性,而不会降低灵敏度。提议的基于MS / MS的ISF筛选也大大减少了无法验证的ISF识别机会。MetaboKit的定量模块产生的峰面积值与在MS1和MS2水平上的代谢物标准品的DIA分析中的已知浓度高度相关。而且,分析 s定量模块产生的峰面积值与DIA标准品和MS2含量水平的代谢物标准品的DIA分析高度相关。而且,分析 s定量模块产生的峰面积值与DIA标准品和MS2含量水平的代谢物标准品的DIA分析高度相关。而且,分析Cdk1 Liv-/-小鼠肝脏表明MetaboKit可以识别多种脂质种类及其ISF,并定量重建这些小鼠中特征明确的脂肪肝表型。在DIA数据中,MS1级和MS2级峰面积数据在微分丰度分析中产生了相似的倍数变化估计值,并且MS2级峰面积数据允许对前体离子色谱图太嘈杂而无法进行峰积分的化合物进行定量比较。
更新日期:2020-06-10
中文翻译:

MetaboKit:用于非目标代谢组学的全面数据提取工具。
我们已经开发了MetaboKit,这是用于基于质谱的非目标代谢组学分析中化合物鉴定和相对定量的综合软件包。在依赖数据的数据采集(DDA)分析中,MetaboKit使用参考光谱库,加合物,二聚体,源内碎片(ISF),MS / MS碎片光谱,以及更重要的是保留时间信息唯一的化合物身份,构建具有化合物身份的定制光谱库实验中使用的色谱系统。使用定制的库,该软件针对DDA分析中的前体离子以及数据独立获取(DIA)分析中的前体离子和产物离子执行目标峰积分。由于其严格的识别算法要求同时匹配MS和MS / MS数据,与竞争性软件包相比,MetaboKit提供的鉴定结果具有更高的特异性,而不会降低灵敏度。提议的基于MS / MS的ISF筛选也大大减少了无法验证的ISF识别机会。MetaboKit的定量模块产生的峰面积值与在MS1和MS2水平上的代谢物标准品的DIA分析中的已知浓度高度相关。而且,分析 s定量模块产生的峰面积值与DIA标准品和MS2含量水平的代谢物标准品的DIA分析高度相关。而且,分析 s定量模块产生的峰面积值与DIA标准品和MS2含量水平的代谢物标准品的DIA分析高度相关。而且,分析Cdk1 Liv-/-小鼠肝脏表明MetaboKit可以识别多种脂质种类及其ISF,并定量重建这些小鼠中特征明确的脂肪肝表型。在DIA数据中,MS1级和MS2级峰面积数据在微分丰度分析中产生了相似的倍数变化估计值,并且MS2级峰面积数据允许对前体离子色谱图太嘈杂而无法进行峰积分的化合物进行定量比较。











































 京公网安备 11010802027423号
京公网安备 11010802027423号