Computer Vision and Image Understanding ( IF 4.3 ) Pub Date : 2020-06-10 , DOI: 10.1016/j.cviu.2020.103009 Daniel Ward , Peyman Moghadam
|
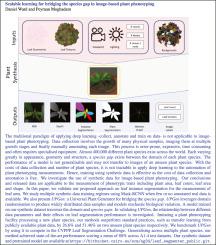
The traditional paradigm of applying deep learning – collect, annotate and train on data – is not applicable to image-based plant phenotyping. Data collection involves the growth of many physical samples, imaging them at multiple growth stages and finally manually annotating each image. This process is error-prone, expensive, time consuming and often requires specialised equipment. Almost 400,000 different plant species exist across the world. Each varying greatly in appearance, geometry and structure, a species gap exists between the domain of each plant species. The performance of a model is not generalisable and may not transfer to images of an unseen plant species. With the costs of data collection and number of plant species, it is not tractable to apply deep learning to the automation of plant phenotyping measurements. Hence, training using synthetic data is effective as the cost of data collection and annotation is free. We investigate the use of synthetic data for image-based plant phenotyping. Our conclusions and released data are applicable to the measurement of phenotypic traits including plant area, leaf count, leaf area and shape. In this paper, we validate our proposed approach on leaf instance segmentation for the measurement of leaf area. We study multiple synthetic data training regimes using Mask-RCNN when few or no annotated real data is available. We also present UPGen: a Universal Plant Generator for bridging the species gap. UPGen leverages domain randomisation to produce widely distributed data samples and models stochastic biological variation. A model trained on our synthetic dataset traverses the domain and species gaps. In validating UPGen, the relationship between different data parameters and their effects on leaf segmentation performance is investigated. Imitating a plant phenotyping facility processing a new plant species, our methods outperform standard practices, such as transfer learning from publicly available plant data, by 26.6% and 51.46% on two unseen plant species respectively. We benchmark UPGen by using it to compete in the CVPPP Leaf Segmentation Challenge. Generalising across multiple plant species, our method achieves state-of-the-art performance scoring a mean of 88% across A1-4 test datasets. Our synthetic dataset and pretrained model are available at https://csiro-robotics.github.io/UPGen-Webpage/.
中文翻译:

基于图像的植物表型可扩展学习弥合物种缺口
应用深度学习的传统范例-收集,注释和培训数据-不适用于基于图像的植物表型。数据收集涉及许多物理样本的生长,在多个生长阶段对它们进行成像,最后手动注释每个图像。该过程容易出错,昂贵,费时并且经常需要专用设备。全世界有近40万种植物。每个在外观,几何形状和结构,大大改变物种间隙存在于每个植物物种的域之间。模型的性能不可概括,并且可能无法转移到看不见的植物物种的图像上。由于数据收集的成本和植物种类的数量,将深度学习应用于植物表型测量的自动化并非易事。因此,使用合成数据进行培训是有效的,因为数据收集和注释的成本是免费的。我们调查基于图像的植物表型的合成数据的使用。我们的结论和公开的数据适用于表型性状的测量,包括植物面积,叶数,叶面积和形状。在本文中,我们验证了我们提出的叶实例分割方法用于测量叶面积。当几乎没有注释的实际数据可用时,我们使用Mask-RCNN研究多种综合数据训练方案。我们还介绍了UPGen:一种通用的植物生成器,用于弥合物种缺口。UPGen利用域随机化来产生分布广泛的数据样本,并对随机生物变异进行建模。在我们的综合数据集上训练的模型可以遍历域和物种之间的差距。在验证UPGen时,研究了不同数据参数之间的关系及其对叶片分割性能的影响。模仿处理新植物物种的植物表型设施,我们的方法优于标准做法,例如从公开的植物数据中转移学习,对两种未见见闻的植物物种分别增加了26.6%和51.46%。我们以UPGen为基准通过使用它来竞争CVPPP叶分割挑战。通过对多种植物进行概括,我们的方法在A1-4测试数据集上实现了平均88%的最新性能得分。我们的综合数据集和预训练模型可在https://csiro-robotics.github.io/UPGen-Webpage/上获得。











































 京公网安备 11010802027423号
京公网安备 11010802027423号