当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Scop3P: A Comprehensive Resource of Human Phosphosites within Their Full Context.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-06-08 , DOI: 10.1021/acs.jproteome.0c00306 Pathmanaban Ramasamy 1, 2, 3, 4, 5 , Demet Turan 1, 2 , Natalia Tichshenko 1, 2 , Niels Hulstaert 1, 2 , Elien Vandermarliere 1, 2 , Wim Vranken 3, 4, 5 , Lennart Martens 1, 2
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-06-08 , DOI: 10.1021/acs.jproteome.0c00306 Pathmanaban Ramasamy 1, 2, 3, 4, 5 , Demet Turan 1, 2 , Natalia Tichshenko 1, 2 , Niels Hulstaert 1, 2 , Elien Vandermarliere 1, 2 , Wim Vranken 3, 4, 5 , Lennart Martens 1, 2
Affiliation
|
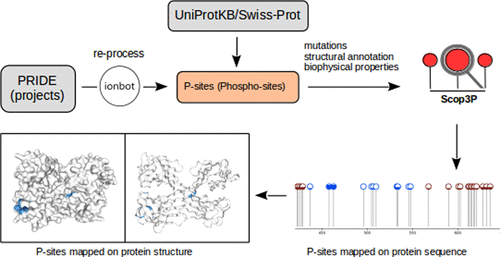
Protein phosphorylation is a key post-translational modification in many biological processes and is associated to human diseases such as cancer and metabolic disorders. The accurate identification, annotation, and functional analysis of phosphosites are therefore crucial to understand their various roles. Phosphosites are mainly analyzed through phosphoproteomics, which has led to increasing amounts of publicly available phosphoproteomics data. Several resources have been built around the resulting phosphosite information, but these are usually restricted to the protein sequence and basic site metadata. What is often missing from these resources, however, is context, including protein structure mapping, experimental provenance information, and biophysical predictions. We therefore developed Scop3P: a comprehensive database of human phosphosites within their full context. Scop3P integrates sequences (UniProtKB/Swiss-Prot), structures (PDB), and uniformly reprocessed phosphoproteomics data (PRIDE) to annotate all known human phosphosites. Furthermore, these sites are put into biophysical context by annotating each phosphoprotein with per-residue structural propensity, solvent accessibility, disordered probability, and early folding information. Scop3P, available at https://iomics.ugent.be/scop3p, presents a unique resource for visualization and analysis of phosphosites and for understanding of phosphosite structure–function relationships.
中文翻译:

Scop3P:全面了解人类磷酸的综合资源。
蛋白质磷酸化是许多生物学过程中的关键翻译后修饰,并与人类疾病如癌症和代谢异常有关。因此,正确识别,注释和进行磷酸位点的功能分析对于理解其各种作用至关重要。主要通过磷酸化蛋白质组学对磷酸酯进行分析,这导致了越来越多的公开可用的磷酸化蛋白质组学数据。已围绕所得的磷酸位点信息建立了几种资源,但这些资源通常仅限于蛋白质序列和基本位点元数据。但是,这些资源通常缺少上下文,包括蛋白质结构图,实验来源信息和生物物理预测。因此,我们开发了Scop3P:全面了解人类磷酸酯的全面数据库。Scop3P整合了序列(UniProtKB / Swiss-Prot),结构(PDB)和统一重新处理的磷酸蛋白质组学数据(PRIDE),以注释所有已知的人类磷酸位点。此外,通过用每个残基的结构倾向性,溶剂可及性,无序概率和早期折叠信息注释每个磷蛋白,将这些位点置于生物物理环境中。Scop3P可在https://iomics.ugent.be/scop3p上获得,它提供了一个独特的资源,用于可视化和分析磷酸位点以及了解磷酸位点结构与功能之间的关系。通过用每个残基的结构倾向,溶剂可及性,无序概率和早期折叠信息注释每个磷蛋白,将这些位点置于生物物理环境中。Scop3P可在https://iomics.ugent.be/scop3p上获得,它提供了一个独特的资源,用于可视化和分析磷酸位点以及了解磷酸位点结构与功能之间的关系。通过用每个残基的结构倾向,溶剂可及性,无序概率和早期折叠信息注释每个磷蛋白,将这些位点置于生物物理环境中。Scop3P可在https://iomics.ugent.be/scop3p上获得,它提供了一个独特的资源,用于可视化和分析磷酸位点以及了解磷酸位点结构与功能之间的关系。
更新日期:2020-08-08
中文翻译:

Scop3P:全面了解人类磷酸的综合资源。
蛋白质磷酸化是许多生物学过程中的关键翻译后修饰,并与人类疾病如癌症和代谢异常有关。因此,正确识别,注释和进行磷酸位点的功能分析对于理解其各种作用至关重要。主要通过磷酸化蛋白质组学对磷酸酯进行分析,这导致了越来越多的公开可用的磷酸化蛋白质组学数据。已围绕所得的磷酸位点信息建立了几种资源,但这些资源通常仅限于蛋白质序列和基本位点元数据。但是,这些资源通常缺少上下文,包括蛋白质结构图,实验来源信息和生物物理预测。因此,我们开发了Scop3P:全面了解人类磷酸酯的全面数据库。Scop3P整合了序列(UniProtKB / Swiss-Prot),结构(PDB)和统一重新处理的磷酸蛋白质组学数据(PRIDE),以注释所有已知的人类磷酸位点。此外,通过用每个残基的结构倾向性,溶剂可及性,无序概率和早期折叠信息注释每个磷蛋白,将这些位点置于生物物理环境中。Scop3P可在https://iomics.ugent.be/scop3p上获得,它提供了一个独特的资源,用于可视化和分析磷酸位点以及了解磷酸位点结构与功能之间的关系。通过用每个残基的结构倾向,溶剂可及性,无序概率和早期折叠信息注释每个磷蛋白,将这些位点置于生物物理环境中。Scop3P可在https://iomics.ugent.be/scop3p上获得,它提供了一个独特的资源,用于可视化和分析磷酸位点以及了解磷酸位点结构与功能之间的关系。通过用每个残基的结构倾向,溶剂可及性,无序概率和早期折叠信息注释每个磷蛋白,将这些位点置于生物物理环境中。Scop3P可在https://iomics.ugent.be/scop3p上获得,它提供了一个独特的资源,用于可视化和分析磷酸位点以及了解磷酸位点结构与功能之间的关系。










































 京公网安备 11010802027423号
京公网安备 11010802027423号