Letters in Drug Design & Discovery ( IF 1.2 ) Pub Date : 2020-05-01 , DOI: 10.2174/1570180815666180914162935 Virendra Nath 1 , Rohini Ahuja 1 , Vipin Kumar 1
|
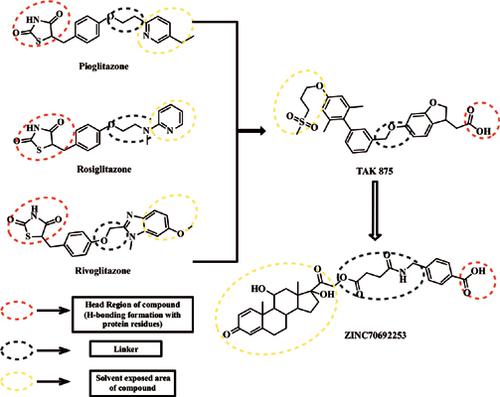
Background: Diabetes is the foremost health problem worldwide predisposing to increased mortality and morbidity. The available synthetic drugs have serious side effects and thus, emphasize further need to develop effective medication therapy. GPR40 represents an interesting target for developing novel antidiabetic drug. In the current study, searching of potential natural hit candidate as agonist by using structure based computational approach.
Methods: The GPR40 agonistic activity of natural compounds was searched by using Maestro through docking and Molecular Dynamics (MD) simulation application. Virtual screening by using IBScreen library of natural compounds was done and the binding modes of newer natural entity(s) were investigated. Further, MD studies of the GPR40 complex with the most promising hit found in this study justified the stability of these complexes.
Results: The silicone chip-based approach recognized the most capable six hits and the ADME prediction aided the exploration of their pharmacokinetic potential. In this study, the obtained hit (ZINC70692253) after the use of exhaustive screening having binding energy -107.501 kcal/mol and root mean square deviation of hGPR40-ZINC70692253 is around 3.5 Å in 20 ns of simulation.
Conclusion: Successful application of structure-based computational screening gave a novel candidate from Natural Product library for diabetes treatment. So, Natural compounds may tend to cure diabetes with lesser extent of undesirable effects in comparison to synthetic compounds and these novel screened compounds may show a plausible biological response in the hit to lead finding of drug development process. To the best of our knowledge, this is the first example of the successful application of SBVS to discover novel natural hit compounds using hGPR40.
中文翻译:

虚拟筛选和计算机模拟分析可快速高效地鉴定新型天然GPR40激动剂
背景:糖尿病是世界范围内最重要的健康问题,容易导致死亡率和发病率的增加。可用的合成药物具有严重的副作用,因此,强调了进一步发展有效药物治疗的需求。GPR40代表开发新型抗糖尿病药物的有趣靶标。在当前的研究中,通过使用基于结构的计算方法来搜索潜在的自然命中候选物作为激动剂。
方法:使用Maestro通过对接和分子动力学(MD)模拟应用程序搜索天然化合物的GPR40激动活性。使用IBScreen天然化合物库进行了虚拟筛选,并研究了新型天然物的结合模式。此外,在这项研究中发现的最有希望的命中的GPR40复合物的MD研究证明了这些复合物的稳定性。
结果:基于有机硅芯片的方法识别出了最有能力的六次点击,而ADME预测则有助于探索其药代动力学潜力。在这项研究中,在20 ns的仿真中,使用具有结合能-107.501 kcal / mol和hGPR40-ZINC70692253的均方根偏差的穷举筛选后获得的命中值(ZINC70692253)为3.5Å。
结论:基于结构的计算筛选的成功应用为天然药物治疗糖尿病患者提供了一个新的候选对象。因此,与合成化合物相比,天然化合物可能会以较小程度的不良影响治愈糖尿病,并且这些新筛选的化合物可能会在药物研发过程中表现出合理的生物学反应。据我们所知,这是成功使用SBVS使用hGPR40发现新型天然命中化合物的第一个例子。











































 京公网安备 11010802027423号
京公网安备 11010802027423号