Current Proteomics ( IF 0.5 ) Pub Date : 2020-05-31 , DOI: 10.2174/1570164617666191004165609 Shihua Chen 1 , Meihu Ma 1 , Xing Fu 1
|
Background: Collagenase is a type of proteolytic enzyme that specifically hydrolyzes native collagen with a three-dimensional helical structure. The structure and properties of collagenase vary with different sources and types. In addition to the well-characterized Clostridium collagenase, other collagenases are largely unknown. Various gene and protein databases have been widely used to mine novel functional genes in the genome. Gene mining and sequence analysis are effective methods for studying these enzymes.
Objective: The present study aimed to understand the molecular, structural, and functional characteristics of collagenase from Bacillus cereus MH19 using a bioinformatics approach.
Methods: Based on the three-generation sequencing PacBio technique, Sequencing the Bacillus cereus MH19 genome. Function annotation is completed by blasting genes with different databases. Collagenases were investigated based on the physiochemical properties, phylogenetic relation, and domain architecture. The 3D structure model of the selected collagenase has been constructed and verified by SAVES.
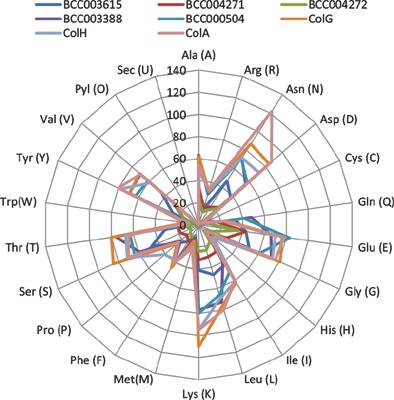
Results: There were 5 collagenases in Bacillus cereus MH19 with a molecular weight distribution ranging from 36-110 kDa. The analysis of evolutionary relationship between different collagenases indicating that the BCC000504 and BCC003388 collagenase gene sequences are closer to the typical collagenase genes ColG, ColA, and ColH, followed by BCC003615. The domain and function analysis showed that the collagenases BCC000504 and BCC003388 were similar to the collagenases ColG, ColA and ColH. BCC004271 was similar to BCC004272, and BCC003615 might be quite different from other collagenases. The secondary structure of collagenase was analyzed. The SAVES evaluation indicates that 3D structural modeling of the selected collagenase is acceptable.
Conclusion: This study provides an overview of the molecular, functional, and structural characteristics of collagenase from Bacillus cereus MH19, which helps to understand the bacterial collagenase. The characterization of the collagenase will certainly expand the application range of collagenase.
中文翻译:

通过计算机方法分析蜡样芽孢杆菌MH19胶原酶的结构和功能特性
背景:胶原酶是一种蛋白水解酶,专门水解具有三维螺旋结构的天然胶原蛋白。胶原酶的结构和性质随来源和类型的不同而变化。除了特征明确的梭菌胶原酶以外,其他胶原酶在很大程度上是未知的。各种基因和蛋白质数据库已被广泛用于挖掘基因组中的新功能基因。基因挖掘和序列分析是研究这些酶的有效方法。
目的:本研究旨在通过生物信息学方法了解蜡状芽孢杆菌MH19胶原酶的分子,结构和功能特性。
方法:基于三代测序PacBio技术,对蜡状芽孢杆菌MH19基因组进行测序。通过使用不同的数据库对基因进行爆炸来完成功能注释。基于生理化学特性,系统发育关系和域结构研究了胶原蛋白。所选胶原酶的3D结构模型已通过SAVES构建并验证。
结果:蜡状芽孢杆菌MH19中有5种胶原酶,分子量分布在36-110 kDa之间。分析不同胶原酶之间的进化关系,表明BCC000504和BCC003388胶原酶基因序列更接近典型的胶原酶基因ColG,ColA和ColH,其次是BCC003615。结构域和功能分析表明,胶原酶BCC000504和BCC003388与胶原酶ColG,ColA和ColH相似。BCC004271与BCC004272相似,并且BCC003615可能与其他胶原酶完全不同。分析了胶原酶的二级结构。SAVES评估表明所选胶原酶的3D结构建模是可以接受的。
结论:本研究概述了蜡样芽孢杆菌MH19胶原酶的分子,功能和结构特征,有助于了解细菌胶原酶。胶原酶的表征必将扩大胶原酶的应用范围。











































 京公网安备 11010802027423号
京公网安备 11010802027423号