Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-02-29 , DOI: 10.2174/1574893614666191202152328 Md. Al Mehedi Hasan 1 , Md Khaled Ben Islam 2 , Julia Rahman 1 , Shamim Ahmad 3
|
Background: Post-translational modification is one of the bio-molecular mechanisms in living organisms, which incorporate functional diversity in proteins as well as regulate cellular processes. Transformation of arginine residue to citrulline in protein is such a modification.
Objective: Our objective is to identify citrullinated arginine residue sites quickly and accurately.
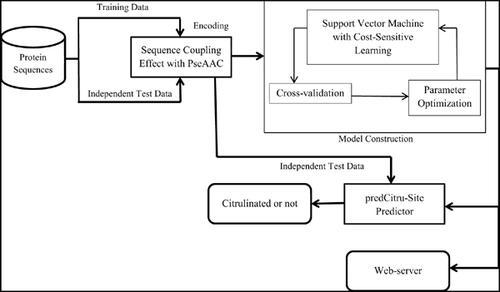
Methods: In this study, a novel computational tool, abbreviated as predCitru-Site, has been developed to predict citrullination sites. This technique effectively has incorporated the sequencecoupling effect of surrounding amino acids of arginine residues as well as optimizes skewed training citrullination dataset for prediction quality improvement. The performance of predCitru- Site has been measured from the average of 5 complete runs of the 10-fold cross-validation test to comply with existing tools.
Results and Conclusion: predCitru-Site has achieved 97.6% sensitivity, 98.9% specificity, and overall accuracy of 98.5%. With Matthew’s correlation coefficient of 0.967, it has also shown an area under the receiver operator characteristics curve of 0.997. Compared with existing tools, predCitru-Site significantly outperforms on the same benchmark dataset. It also shows significant improvement in the case of independent tests in all performance metrics (around 50% higher in AUC). These results suggest that our method is promising and can be used as a complementary technique for fast exploration of citrullination in arginine residue. A user-friendly web server has also been deployed at http://research.ru.ac.bd/predCitru-Site/ for the convenience of experimental scientists.
中文翻译:

通过将序列耦合效应纳入PseAAC并解决数据不平衡问题来预测柠檬酸位点
背景:翻译后修饰是活生物体中的一种生物分子机制,该机制将蛋白质的功能多样性并调节细胞过程。蛋白质中精氨酸残基向瓜氨酸的转化就是这样的修饰。
目的:我们的目标是快速,准确地识别瓜氨酸精氨酸残基。
方法:在这项研究中,已开发出一种新的计算工具,缩写为predCitru-Site,可预测瓜氨酸化位点。该技术有效地结合了精氨酸残基周围氨基酸的序列耦合作用,并优化了偏斜训练瓜氨酸化数据集以提高预测质量。predCitru- Site的性能是根据10倍交叉验证测试的5次完整运行的平均值进行测量的,以符合现有工具的要求。
结果与结论:predCitru-Site的灵敏度为97.6%,特异性为98.9%,总准确度为98.5%。在Matthew的相关系数为0.967的情况下,它还显示出接收器操作员特性曲线下的面积为0.997。与现有工具相比,predCitru-Site在相同基准数据集上的性能明显优于其他工具。在所有性能指标的独立测试中,它也显示出显着的改进(AUC大约提高了50%)。这些结果表明,我们的方法是有前途的,可以用作快速探索精氨酸残基中瓜氨酸化的补充技术。为了方便实验科学家,还在http://research.ru.ac.bd/predCitru-Site/部署了用户友好的Web服务器。











































 京公网安备 11010802027423号
京公网安备 11010802027423号