当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mycoplasma pneumoniae Genome Editing Based on Oligo Recombineering and Cas9-Mediated Counterselection.
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-06-05 , DOI: 10.1021/acssynbio.0c00022 Carlos Piñero-Lambea 1 , Eva Garcia-Ramallo 1 , Sira Martinez 1 , Javier Delgado 1 , Luis Serrano 1, 2 , Maria Lluch-Senar 1, 3
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-06-05 , DOI: 10.1021/acssynbio.0c00022 Carlos Piñero-Lambea 1 , Eva Garcia-Ramallo 1 , Sira Martinez 1 , Javier Delgado 1 , Luis Serrano 1, 2 , Maria Lluch-Senar 1, 3
Affiliation
|
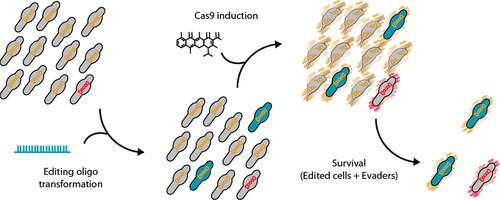
Mycoplasma species share a set of features, such as lack of a cell wall, streamlined genomes, simplified metabolism, and the use of a deviant genetic code, that make them attractive approximations of what a chassis strain should ideally be. Among them, Mycoplasma pneumoniae arises as a candidate for synthetic biology projects, as it is one of the most deeply characterized bacteria. However, the historical paucity of tools for editing Mycoplasma genomes has precluded the establishment of M. pneumoniae as a suitable chassis strain. Here, we developed an oligonucleotide recombineering method for this strain based on GP35, a ssDNA recombinase originally encoded by a Bacillus subtilis-associated phage. GP35-mediated oligo recombineering is able to carry out point mutations in the M. pneumoniae genome with an efficiency as high as 2.7 × 10–2, outperforming oligo recombineering protocols developed for other bacteria. Gene deletions of different sizes showed a decreasing power trend between efficiency and the scale of the attempted edition. However, the editing rates for all modifications increased when CRISPR/Cas9 was used to counterselect nonedited cells. This allowed edited clones carrying chromosomal deletions of up to 1.8 kb to be recovered with little to no screening of survivor cells. We envision this technology as a major step toward the use of M. pneumoniae, and possibly other Mycoplasmas, as synthetic biology chassis strains.
中文翻译:

基于Oligo重组和Cas9介导的反选择的肺炎支原体基因组编辑。
支原体物种具有一系列特征,例如缺少细胞壁,简化的基因组,简化的代谢以及使用异常的遗传密码,这使它们成为理想的底盘菌株的诱人近似物。其中,肺炎支原体是合成生物学项目的候选者,因为它是特征最深的细菌之一。但是,由于历史上缺乏用于编辑支原体基因组的工具,因此无法将肺炎支原体确立为合适的底盘菌株。在这里,我们基于GP35(一种最初由枯草芽孢杆菌编码的ssDNA重组酶)开发了针对该菌株的寡核苷酸重组方法-相关噬菌体。GP35介导的寡聚重组能够在肺炎支原体基因组中进行点突变,效率高达2.7×10 -2,优于其他细菌开发的寡聚重组方案。不同大小的基因缺失显示出效率与尝试版本规模之间的递减功率趋势。然而,当使用CRISPR / Cas9反选择未编辑的细胞时,所有修饰的编辑率均增加。这样就可以在几乎没有筛选幸存细胞的情况下恢复携带高达1.8 kb染色体缺失的编辑克隆。我们设想这项技术是迈入肺炎支原体和其他支原体作为合成生物学底盘菌株的重要一步。
更新日期:2020-07-17
中文翻译:

基于Oligo重组和Cas9介导的反选择的肺炎支原体基因组编辑。
支原体物种具有一系列特征,例如缺少细胞壁,简化的基因组,简化的代谢以及使用异常的遗传密码,这使它们成为理想的底盘菌株的诱人近似物。其中,肺炎支原体是合成生物学项目的候选者,因为它是特征最深的细菌之一。但是,由于历史上缺乏用于编辑支原体基因组的工具,因此无法将肺炎支原体确立为合适的底盘菌株。在这里,我们基于GP35(一种最初由枯草芽孢杆菌编码的ssDNA重组酶)开发了针对该菌株的寡核苷酸重组方法-相关噬菌体。GP35介导的寡聚重组能够在肺炎支原体基因组中进行点突变,效率高达2.7×10 -2,优于其他细菌开发的寡聚重组方案。不同大小的基因缺失显示出效率与尝试版本规模之间的递减功率趋势。然而,当使用CRISPR / Cas9反选择未编辑的细胞时,所有修饰的编辑率均增加。这样就可以在几乎没有筛选幸存细胞的情况下恢复携带高达1.8 kb染色体缺失的编辑克隆。我们设想这项技术是迈入肺炎支原体和其他支原体作为合成生物学底盘菌株的重要一步。









































 京公网安备 11010802027423号
京公网安备 11010802027423号