当前位置:
X-MOL 学术
›
RSC Med. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Systematic analysis of the interactions driving small molecule–RNA recognition
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2020-06-04 , DOI: 10.1039/d0md00167h G Padroni 1 , N N Patwardhan 1 , M Schapira 2, 3 , A E Hargrove 1
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2020-06-04 , DOI: 10.1039/d0md00167h G Padroni 1 , N N Patwardhan 1 , M Schapira 2, 3 , A E Hargrove 1
Affiliation
|
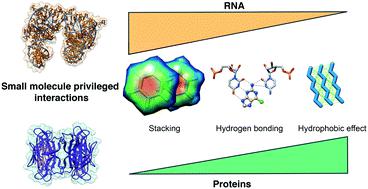
RNA molecules are becoming an important target class in drug discovery. However, the principles for designing RNA-binding small molecules are yet to be fully uncovered. In this study, we examined the Protein Data Bank (PDB) to highlight privileged interactions underlying small molecule–RNA recognition. By comparing this analysis with previously determined small molecule–protein interactions, we find that RNA recognition is driven mostly by stacking and hydrogen bonding interactions, while protein recognition is instead driven by hydrophobic effects. Furthermore, we analyze patterns of interactions to highlight potential strategies to tune RNA recognition, such as stacking and cation–π interactions that favor purine and guanine recognition, and note an unexpected paucity of backbone interactions, even for cationic ligands. Collectively, this work provides further understanding of RNA–small molecule interactions that may inform the design of small molecules targeting RNA.
中文翻译:

驱动小分子-RNA识别的相互作用的系统分析
RNA 分子正在成为药物发现的重要目标类别。然而,设计RNA结合小分子的原理尚未完全揭示。在这项研究中,我们检查了蛋白质数据库 (PDB),以突出显示小分子 - RNA 识别的特权相互作用。通过将此分析与先前确定的小分子-蛋白质相互作用进行比较,我们发现 RNA 识别主要由堆叠和氢键相互作用驱动,而蛋白质识别则由疏水效应驱动。此外,我们分析了相互作用的模式以突出调整 RNA 识别的潜在策略,例如有利于嘌呤和鸟嘌呤识别的堆叠和阳离子-π 相互作用,并注意到骨架相互作用的意外缺乏,即使对于阳离子配体也是如此。集体,
更新日期:2020-07-22
中文翻译:

驱动小分子-RNA识别的相互作用的系统分析
RNA 分子正在成为药物发现的重要目标类别。然而,设计RNA结合小分子的原理尚未完全揭示。在这项研究中,我们检查了蛋白质数据库 (PDB),以突出显示小分子 - RNA 识别的特权相互作用。通过将此分析与先前确定的小分子-蛋白质相互作用进行比较,我们发现 RNA 识别主要由堆叠和氢键相互作用驱动,而蛋白质识别则由疏水效应驱动。此外,我们分析了相互作用的模式以突出调整 RNA 识别的潜在策略,例如有利于嘌呤和鸟嘌呤识别的堆叠和阳离子-π 相互作用,并注意到骨架相互作用的意外缺乏,即使对于阳离子配体也是如此。集体,











































 京公网安备 11010802027423号
京公网安备 11010802027423号