当前位置:
X-MOL 学术
›
Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Site-Specific Integration Reporter System That Enables Rapid Evaluation of CRISPR/Cas9-Mediated Genome Editing Strategies in CHO Cells.
Biotechnology Journal ( IF 3.2 ) Pub Date : 2020-06-04 , DOI: 10.1002/biot.202000057 Nathaniel K Hamaker 1, 2 , Kelvin H Lee 1, 2
Biotechnology Journal ( IF 3.2 ) Pub Date : 2020-06-04 , DOI: 10.1002/biot.202000057 Nathaniel K Hamaker 1, 2 , Kelvin H Lee 1, 2
Affiliation
|
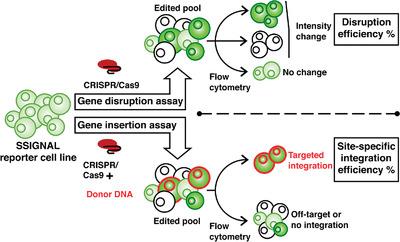
Targeted gene knockout and site‐specific integration (SSI) are powerful genome editing techniques to improve the development of industrially relevant Chinese hamster ovary (CHO) cell lines. However, past efforts to perform SSI in CHO cells are characterized by low efficiencies. Moreover, numerous strategies proposed to boost SSI efficiency in mammalian cell types have yet to be evaluated head to head or in combination to appreciably boost efficiencies in CHO. To enable systematic and rapid optimization of genome editing methods, the SSIGNAL (site‐specific integration and genome alteration) reporter system is developed. This tool can analyze CRISPR (clustered regularly interspaced palindromic repeats)/Cas9 (CRISPR‐associated protein 9)‐mediated disruption activity alone or in conjunction with SSI efficiency. The reporter system uses green and red dual‐fluorescence signals to indicate genotype states within four days following transfection, facilitating rapid data acquisition via standard flow cytometry instrumentation. In addition to describing the design and development of the system, two of its applications are demonstrated by first comparing transfection conditions to maximize CRISPR/Cas9 activity and subsequently assessing the efficiency of several promising SSI strategies. Due to its sensitivity and versatility, the SSIGNAL reporter system may serve as a tool to advance genome editing technology.
更新日期:2020-08-04











































 京公网安备 11010802027423号
京公网安备 11010802027423号