当前位置:
X-MOL 学术
›
J. Med. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Computer Vision Approach to Align and Compare Protein Cavities: Application to Fragment-Based Drug Design.
Journal of Medicinal Chemistry ( IF 6.8 ) Pub Date : 2020-06-04 , DOI: 10.1021/acs.jmedchem.0c00422 Merveille Eguida 1 , Didier Rognan 1
Journal of Medicinal Chemistry ( IF 6.8 ) Pub Date : 2020-06-04 , DOI: 10.1021/acs.jmedchem.0c00422 Merveille Eguida 1 , Didier Rognan 1
Affiliation
|
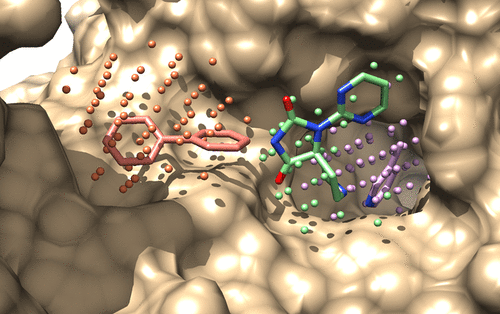
Identifying local similarities in binding sites from distant proteins is a major hurdle to rational drug design. We herewith present a novel method, borrowed from computer vision, adapted to mine fragment subpockets and compare them to whole ligand-binding sites. Pockets are represented by pharmacophore-annotated point clouds mimicking ideal ligands or fragments. Point cloud registration is used to find the transformation enabling an optimal overlap of points sharing similar topological and pharmacophoric neighborhoods. The method (ProCare) was calibrated on a large set of druggable cavities and applied to the comparison of fragment subpockets to entire cavities. A collection of 33,953 subpockets annotated with their bound fragments was screened for local similarity to cavities from recently described protein X-ray structures. ProCare was able to detect local similarities between remote pockets and transfer the corresponding fragments to the query cavity space, thereby proposing a first step to fragment-based design approaches targeting orphan cavities.
中文翻译:

对齐和比较蛋白质腔的计算机视觉方法:在基于片段的药物设计中的应用。
鉴定远距离蛋白质结合位点的局部相似性是合理药物设计的主要障碍。我们在此提出一种借鉴计算机视觉的新颖方法,该方法适用于挖掘碎片子口袋并将它们与整个配体结合位点进行比较。口袋由模仿理想配体或片段的药效基团注释的点云表示。点云配准用于查找转换,以实现共享相似拓扑和药效比邻域的点的最佳重叠。该方法(ProCare)在大量可药物化的腔体上进行了校准,并用于比较子腔与整个腔体的比较。从最近描述的蛋白质X射线结构中筛选了33,953个子兜的集合,这些子兜用其结合的片段进行注释,以查找与空腔的局部相似性。
更新日期:2020-07-09
中文翻译:

对齐和比较蛋白质腔的计算机视觉方法:在基于片段的药物设计中的应用。
鉴定远距离蛋白质结合位点的局部相似性是合理药物设计的主要障碍。我们在此提出一种借鉴计算机视觉的新颖方法,该方法适用于挖掘碎片子口袋并将它们与整个配体结合位点进行比较。口袋由模仿理想配体或片段的药效基团注释的点云表示。点云配准用于查找转换,以实现共享相似拓扑和药效比邻域的点的最佳重叠。该方法(ProCare)在大量可药物化的腔体上进行了校准,并用于比较子腔与整个腔体的比较。从最近描述的蛋白质X射线结构中筛选了33,953个子兜的集合,这些子兜用其结合的片段进行注释,以查找与空腔的局部相似性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号