Cell ( IF 45.5 ) Pub Date : 2020-06-04 , DOI: 10.1016/j.cell.2020.04.056 J Brooks Crickard 1 , Corentin J Moevus 1 , Youngho Kwon 2 , Patrick Sung 2 , Eric C Greene 1
|
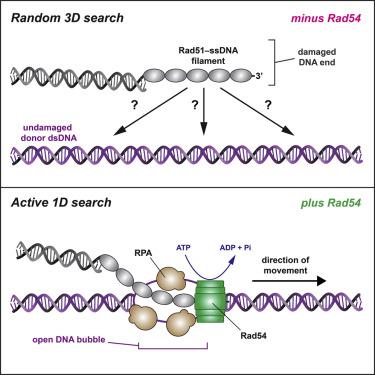
Homologous recombination (HR) helps maintain genome integrity, and HR defects give rise to disease, especially cancer. During HR, damaged DNA must be aligned with an undamaged template through a process referred to as the homology search. Despite decades of study, key aspects of this search remain undefined. Here, we use single-molecule imaging to demonstrate that Rad54, a conserved Snf2-like protein found in all eukaryotes, switches the search from the diffusion-based pathways characteristic of the basal HR machinery to an active process in which DNA sequences are aligned via an ATP-dependent molecular motor-driven mechanism. We further demonstrate that Rad54 disrupts the donor template strands, enabling the search to take place within a migrating DNA bubble-like structure that is bound by replication protein A (RPA). Our results reveal that Rad54, working together with RPA, fundamentally alters how DNA sequences are aligned during HR.
中文翻译:

Rad54在同源重组过程中驱动ATP水解相关的DNA序列比对。
同源重组(HR)有助于维持基因组完整性,并且HR缺陷会导致疾病,尤其是癌症。在HR期间,必须通过称为同源性搜索的过程将受损的DNA与未损坏的模板对齐。尽管进行了数十年的研究,但此搜索的关键方面仍不确定。在这里,我们使用单分子成像来证明Rad54(一种在所有真核生物中均发现的保守Snf2样蛋白)将搜索从基础HR机械的基于扩散的途径转换为一个主动过程,在该过程中DNA序列通过ATP依赖性分子马达驱动的机制。我们进一步证明,Rad54破坏供体模板链,使搜索能够在由复制蛋白A(RPA)结合的DNA气泡状迁移结构中进行。











































 京公网安备 11010802027423号
京公网安备 11010802027423号