Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2020-06-02 , DOI: 10.1016/j.csbj.2020.05.024 Alireza Sahaf Naeini 1, 2 , Amna Farooq 1 , Magnar Bjørås 2, 3 , Junbai Wang 1
|
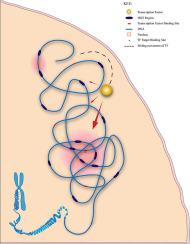
The human genome is regulated in a multi-dimensional way. While biophysical factors like Non-specific Transcription factor Binding Affinity (nTBA) act at DNA sequence level, other factors act above sequence levels such as histone modifications and 3-D chromosomal interactions. This multidimensionality of regulation requires many of these factors for a proper understanding of the regulatory landscape of the human genome. Here, we propose a new biophysical model for estimating nTBA. Integration of nTBA with chromatin modifications and chromosomal interactions, using a new Integrative Genome Analysis Pipeline (IGAP), reveals additive effects of nTBA to regulatory DNA sequences and identifies three types of genomic zones in the human genome (Inactive Genomic Zones, Poised Genomic Zones, and Active Genomic Zones). It also unveils a novel long distance gene regulatory model: chromosomal interactions reduce the physical distance between the high occupancy target (HOT) regions that results in high nTBA to DNA in the area, which in turn attract TFs to such regions with higher binding potential. These findings will help to elucidate the three-dimensional diffusion process that TFs use during their search for the right targets.
中文翻译:

IGAP整合基因组分析流程揭示了与非特异性TF-DNA结合亲和力相关的新基因调控模型。
人类基因组以多维方式调控。虽然诸如非特异性转录因子结合亲和力(nTBA)之类的生物物理因子在DNA序列水平上起作用,但其他因子在序列水平之上也起作用,例如组蛋白修饰和3-D染色体相互作用。这种调节的多维性需要许多这些因素才能正确理解人类基因组的调节态势。在这里,我们提出了一个新的生物物理模型来估算nTBA。使用新的整合基因组分析管道(IGAP),将nTBA与染色质修饰和染色体相互作用整合在一起,揭示了nTBA对调控DNA序列的累加效应,并鉴定了人类基因组中的三种类型的基因组区域(无效基因组区域,平衡基因组区域,和有效基因组区)。它还揭示了一种新颖的长距离基因调控模型:染色体相互作用减少了高占有率目标(HOT)区之间的物理距离,从而导致该区域DNA的nTBA高,进而将TF吸引到具有更高结合潜力的此类区域。这些发现将有助于阐明TF在寻找正确目标时使用的三维扩散过程。











































 京公网安备 11010802027423号
京公网安备 11010802027423号