当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Enzyme-Substrate-Cofactor Dynamical Networks Revealed by High-Resolution Field Cycling Relaxometry.
Biochemistry ( IF 2.9 ) Pub Date : 2020-06-01 , DOI: 10.1021/acs.biochem.0c00212 Masha M Rosenberg 1 , Tianjiong Yao 1 , Gregory C Patton 1 , Alfred G Redfield 2 , Mary F Roberts 3 , Lizbeth Hedstrom 1, 4
Biochemistry ( IF 2.9 ) Pub Date : 2020-06-01 , DOI: 10.1021/acs.biochem.0c00212 Masha M Rosenberg 1 , Tianjiong Yao 1 , Gregory C Patton 1 , Alfred G Redfield 2 , Mary F Roberts 3 , Lizbeth Hedstrom 1, 4
Affiliation
|
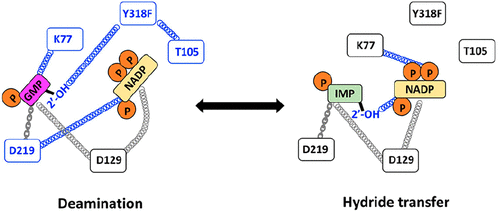
The remarkable power and specificity of enzyme catalysis rely on the dynamic alignment of the enzyme, substrates, and cofactors, yet the role of dynamics has usually been approached from the perspective of the protein. We have been using an underappreciated NMR technique, subtesla high-resolution field cycling 31P NMR relaxometry, to investigate the dynamics of the enzyme-bound substrates and cofactor on guanosine-5′-monophosphate reductase (GMPR). GMPR forms two dead end, yet catalytically competent, complexes that mimic distinct steps in the catalytic cycle: E·IMP·NADP+ undergoes a partial hydride transfer reaction, while E·GMP·NADP+ undergoes a partial deamination reaction. A different cofactor conformation is required for each partial reaction. Here we report the effects of mutations designed to perturb cofactor conformation and ammonia binding with the goal of identifying the structural features that contribute to the distinct dynamic signatures of the hydride transfer and deamination complexes. These experiments suggest that Asp129 is a central cog in a dynamic network required for both hydride transfer and deamination. In contrast, Lys77 modulates the conformation and mobility of substrates and cofactors in a reaction-specific manner. Thr105 and Tyr318 are part of a deamination-specific dynamic network that includes the 2′-OH of GMP. These residues have comparatively little effect on the dynamic properties of the hydride transfer complex. These results further illustrate the potential of high-resolution field cycling NMR relaxometry for the investigation of ligand dynamics. In addition, exchange experiments indicate that NH3/NH4+ has a high affinity for the deamination complex but a low affinity for the hydride transfer complex, suggesting that the movement of ammonia may gate the cofactor conformational change. Collectively, these experiments reinforce the view that the enzyme, substrates, and cofactor are linked in intricate, reaction-specific, dynamic networks and demonstrate that distal portions of the substrates and cofactors are critical features in these networks.
中文翻译:

高分辨率场循环松弛测量揭示的酶-底物-辅因子动态网络。
酶催化的卓越能力和特异性依赖于酶、底物和辅因子的动态排列,但动力学的作用通常是从蛋白质的角度来探讨的。我们一直在使用一种未被充分重视的 NMR 技术,即亚特斯拉高分辨率场循环31 P NMR 弛豫测定法,来研究 5'-单磷酸鸟苷还原酶 (GMPR) 上的酶结合底物和辅因子的动力学。 GMPR 形成两个死胡同,但具有催化能力,复合物模拟催化循环中的不同步骤:E·IMP·NADP +经历部分氢化物转移反应,而 E·GMP·NADP +经历部分脱氨反应。每个部分反应需要不同的辅因子构象。在这里,我们报告了旨在扰乱辅因子构象和氨结合的突变的影响,目的是确定有助于氢化物转移和脱氨复合物的独特动态特征的结构特征。这些实验表明,Asp129 是氢化物转移和脱氨所需的动态网络中的中心齿轮。相比之下,Lys77 以反应特异性方式调节底物和辅因子的构象和迁移率。 Thr105 和 Tyr318 是脱氨特异性动态网络的一部分,其中包括 GMP 的 2'-OH。这些残基对氢化物转移配合物的动态性质影响相对较小。这些结果进一步说明了高分辨率场循环核磁共振弛豫测量在配体动力学研究中的潜力。 此外,交换实验表明NH 3 /NH 4 +对脱氨复合物具有高亲和力,但对氢化物转移复合物具有低亲和力,这表明氨的运动可能控制辅助因子构象变化。总的来说,这些实验强化了酶、底物和辅因子在复杂的、反应特异性的动态网络中相互连接的观点,并证明底物和辅因子的远端部分是这些网络中的关键特征。
更新日期:2020-06-30
中文翻译:

高分辨率场循环松弛测量揭示的酶-底物-辅因子动态网络。
酶催化的卓越能力和特异性依赖于酶、底物和辅因子的动态排列,但动力学的作用通常是从蛋白质的角度来探讨的。我们一直在使用一种未被充分重视的 NMR 技术,即亚特斯拉高分辨率场循环31 P NMR 弛豫测定法,来研究 5'-单磷酸鸟苷还原酶 (GMPR) 上的酶结合底物和辅因子的动力学。 GMPR 形成两个死胡同,但具有催化能力,复合物模拟催化循环中的不同步骤:E·IMP·NADP +经历部分氢化物转移反应,而 E·GMP·NADP +经历部分脱氨反应。每个部分反应需要不同的辅因子构象。在这里,我们报告了旨在扰乱辅因子构象和氨结合的突变的影响,目的是确定有助于氢化物转移和脱氨复合物的独特动态特征的结构特征。这些实验表明,Asp129 是氢化物转移和脱氨所需的动态网络中的中心齿轮。相比之下,Lys77 以反应特异性方式调节底物和辅因子的构象和迁移率。 Thr105 和 Tyr318 是脱氨特异性动态网络的一部分,其中包括 GMP 的 2'-OH。这些残基对氢化物转移配合物的动态性质影响相对较小。这些结果进一步说明了高分辨率场循环核磁共振弛豫测量在配体动力学研究中的潜力。 此外,交换实验表明NH 3 /NH 4 +对脱氨复合物具有高亲和力,但对氢化物转移复合物具有低亲和力,这表明氨的运动可能控制辅助因子构象变化。总的来说,这些实验强化了酶、底物和辅因子在复杂的、反应特异性的动态网络中相互连接的观点,并证明底物和辅因子的远端部分是这些网络中的关键特征。











































 京公网安备 11010802027423号
京公网安备 11010802027423号