当前位置:
X-MOL 学术
›
Mol. Pharmaceutics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Direct Comparison of Total Clearance Prediction: Computational Machine Learning Model versus Bottom-Up Approach Using In Vitro Assay.
Molecular Pharmaceutics ( IF 4.5 ) Pub Date : 2020-06-01 , DOI: 10.1021/acs.molpharmaceut.9b01294 Yohei Kosugi 1 , Natalie Hosea 1
Molecular Pharmaceutics ( IF 4.5 ) Pub Date : 2020-06-01 , DOI: 10.1021/acs.molpharmaceut.9b01294 Yohei Kosugi 1 , Natalie Hosea 1
Affiliation
|
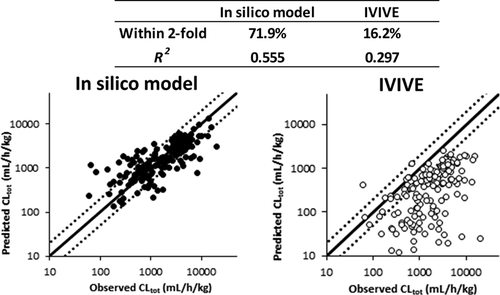
The in vitro–in vivo extrapolation (IVIVE) approach for predicting total plasma clearance (CLtot) has been widely used to rank order compounds early in discovery. More recently, a computational machine learning approach utilizing physicochemical descriptors and fingerprints calculated from chemical structure information has emerged, enabling virtual predictions even earlier in discovery. Previously, this approach focused more on in vitro intrinsic clearance (CLint) prediction. Herein, we directly compare these two approaches for predicting CLtot in rats. A structurally diverse set of 1114 compounds with known in vivo CLtot, in vitro CLint, and plasma protein binding was used as the basis for this evaluation. The machine learning models were assessed by validation approaches using the time- and cluster-split training and test sets, and five-fold cross validation. Assessed by five-fold validation, the random forest regression (RF) and radial basis function (RBF) models demonstrated better prediction performance in eight attempted machine learning models. The CLtot values predicted by the RF and RBF models were within two-fold of the observed values for 67.7 and 71.9% of cluster-split test set compounds, respectively, while the predictivity was worse in the time-split dataset. The predictivity of both models tended to be improved by incorporating in vitro parameters, unbound fraction in plasma (fu,p), and CLint. CLtot prediction utilizing in vitro CLint and the well-stirred model, correcting for the fraction unbound in blood, was substantially worse compared to machine learning approaches for the same cluster-split test set. The reason that CLtot is underestimated by IVIVE is not fully explained by considering the calculated microsomal unbound fraction (cfu,mic), extended clearance classification system (ECCS), and omitting high clearance compounds in excess of hepatic blood flow. The analysis suggests that in silico machine learning models may have the power to reduce reliance on or replace in vitro and in vivo studies for chemical structure optimization in early drug discovery.
中文翻译:

总清除率预测的直接比较:使用体外分析的计算式机器学习模型与自下而上的方法。
用于预测总血浆清除率(CL tot)的体外-体内外推(IVIVE)方法已被广泛用于在发现早期对有序化合物进行排名。最近,出现了一种利用物理化学描述符和从化学结构信息计算出的指纹的计算机学习方法,甚至可以在发现的更早阶段进行虚拟预测。以前,这种方法更多地侧重于体外固有清除率(CL int)预测。在这里,我们直接比较这两种预测大鼠CL tot的方法。结构多样的1114种化合物,具有已知的体内CL tot和体外CL int,并且血浆蛋白结合被用作该评估的基础。通过使用时间和群集拆分训练和测试集以及五重交叉验证的验证方法来评估机器学习模型。通过五重验证进行评估,随机森林回归(RF)和径向基函数(RBF)模型在八种尝试的机器学习模型中表现出更好的预测性能。通过RF和RBF模型预测的CL tot值分别是对集群拆分测试集化合物的67.7%和71.9%的观察值的两倍,而在时间拆分数据集中,可预测性更差。通过合并体外参数,血浆中未结合分数(f u,p)和CL ,两种模型的预测性趋于提高。INT。与针对同一集群拆分测试集的机器学习方法相比,利用体外CL int和良好搅拌的模型校正血液中未结合部分的CL tot预测要差得多。通过考虑计算的微粒体未结合分数(cf u,mic),扩展清除分类系统(ECCS)并忽略肝血流过多的高清除率化合物,无法充分解释IVIVE对CL tot的低估原因。分析表明,计算机模拟机器学习模型可能会减少对早期药物发现中化学结构优化的依赖或替代体外和体内研究。
更新日期:2020-07-06
中文翻译:

总清除率预测的直接比较:使用体外分析的计算式机器学习模型与自下而上的方法。
用于预测总血浆清除率(CL tot)的体外-体内外推(IVIVE)方法已被广泛用于在发现早期对有序化合物进行排名。最近,出现了一种利用物理化学描述符和从化学结构信息计算出的指纹的计算机学习方法,甚至可以在发现的更早阶段进行虚拟预测。以前,这种方法更多地侧重于体外固有清除率(CL int)预测。在这里,我们直接比较这两种预测大鼠CL tot的方法。结构多样的1114种化合物,具有已知的体内CL tot和体外CL int,并且血浆蛋白结合被用作该评估的基础。通过使用时间和群集拆分训练和测试集以及五重交叉验证的验证方法来评估机器学习模型。通过五重验证进行评估,随机森林回归(RF)和径向基函数(RBF)模型在八种尝试的机器学习模型中表现出更好的预测性能。通过RF和RBF模型预测的CL tot值分别是对集群拆分测试集化合物的67.7%和71.9%的观察值的两倍,而在时间拆分数据集中,可预测性更差。通过合并体外参数,血浆中未结合分数(f u,p)和CL ,两种模型的预测性趋于提高。INT。与针对同一集群拆分测试集的机器学习方法相比,利用体外CL int和良好搅拌的模型校正血液中未结合部分的CL tot预测要差得多。通过考虑计算的微粒体未结合分数(cf u,mic),扩展清除分类系统(ECCS)并忽略肝血流过多的高清除率化合物,无法充分解释IVIVE对CL tot的低估原因。分析表明,计算机模拟机器学习模型可能会减少对早期药物发现中化学结构优化的依赖或替代体外和体内研究。











































 京公网安备 11010802027423号
京公网安备 11010802027423号