Journal of Biomedical informatics ( IF 4.0 ) Pub Date : 2020-06-01 , DOI: 10.1016/j.jbi.2020.103455 Jeya Balaji Balasubramanian 1 , Rebecca D Boes 2 , Vanathi Gopalakrishnan 3
|
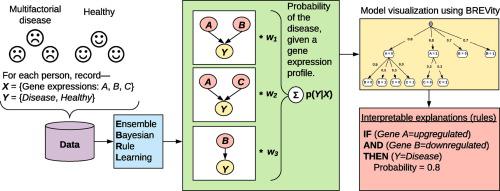
Modeling factors influencing disease phenotypes, from biomarker profiling study datasets, is a critical task in biomedicine. Such datasets are typically generated from high-throughput ’omic’ technologies, which help examine disease mechanisms at an unprecedented resolution. These datasets are challenging because they are high-dimensional. The disease mechanisms they study are also complex because many diseases are multifactorial, resulting from the collective activity of several factors, each with a small effect. Bayesian rule learning (BRL) is a rule model inferred from learning Bayesian networks from data, and has been shown to be effective in modeling high-dimensional datasets. However, BRL is not efficient at modeling multifactorial diseases since it suffers from data fragmentation during learning. In this paper, we overcome this limitation by implementing and evaluating three types of ensemble model combination strategies with BRL— uniform combination (UC; same as Bagging), Bayesian model averaging (BMA), and Bayesian model combination (BMC)— collectively called Ensemble Bayesian Rule Learning (EBRL). We also introduce a novel method to visualize EBRL models, called the Bayesian Rule Ensemble Visualizing tool (BREVity), which helps extract interpret the most important rule patterns guiding the predictions made by the ensemble model. Our results using twenty-five public, high-dimensional, gene expression datasets of multifactorial diseases, suggest that, both EBRL models using UC and BMC achieve better predictive performance than BMA and other classic machine learning methods. Furthermore, BMC is found to be more reliable than UC, when the ensemble includes sub-optimal models resulting from the stochasticity of the model search process. Together, EBRL and BREVity provides researchers a promising and novel tool for modeling multifactorial diseases from high-dimensional datasets that leverages strengths of ensemble methods for predictive performance, while also providing interpretable explanations for its predictions.
中文翻译:

一种使用Ensemble贝叶斯规则分类器对多因素疾病进行建模的新颖方法。
来自生物标志物分析研究数据集的影响疾病表型的建模因素是生物医学中的关键任务。此类数据集通常是通过高通量“组学”技术生成的,可帮助以前所未有的分辨率检查疾病机制。这些数据集具有较高的维数,因此具有挑战性。他们研究的疾病机制也很复杂,因为许多疾病是多种因素造成的,这是由多种因素的共同活动导致的,每种因素影响很小。贝叶斯规则学习(BRL)是从数据中学习贝叶斯网络推断出的规则模型,并已证明在建模高维数据集方面有效。但是,BRL在建模多因素疾病方面效率不高,因为它在学习过程中会遭受数据碎片的困扰。在本文中,我们通过使用BRL实施和评估三种类型的集成模型组合策略来克服此限制-统一组合(UC;与Bagging相同),贝叶斯模型平均(BMA)和贝叶斯模型组合(BMC)-统称为Ensemble贝叶斯规则学习( EBRL)。我们还介绍了一种新的可视化EBRL模型的方法,称为贝叶斯规则集成可视化工具(BREVity),该方法可帮助提取解释最重要的规则模式的方法,这些规则模式指导了集成模型的预测。我们使用25个多因素疾病公共,高维基因表达数据集的结果表明,使用UC和BMC的EBRL模型均比BMA和其他经典的机器学习方法具有更好的预测性能。此外,发现BMC比UC更可靠,当集合中包含由模型搜索过程的随机性导致的次优模型时。EBRL和BREVity共同为研究人员提供了一种有前途的新颖工具,可用于从高数据集建模多因子疾病,该模型利用集成方法的优势来实现预测性能,同时也为其预测提供可解释的解释。










































 京公网安备 11010802027423号
京公网安备 11010802027423号