Molecular Phylogenetics and Evolution ( IF 3.6 ) Pub Date : 2020-05-28 , DOI: 10.1016/j.ympev.2020.106857 Lingfeng Kong 1 , Yuanning Li 2 , Kevin M Kocot 3 , Yi Yang 4 , Lu Qi 4 , Qi Li 1 , Kenneth M Halanych 5
|
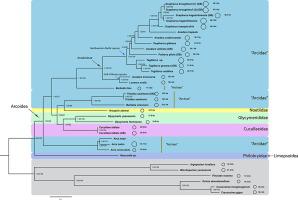
Arcoida, comprising about 570 species of blood cockles, is an ecologically and economically important lineage of bivalve molluscs. Current classification of arcoids is largely based on morphology, which shows widespread homoplasy. Despite two recent studies employing multi-locus analyses with broad sampling of Arcoida, evolutionary relationships among major lineages remain controversial. Interestingly, mitochondrial genomes of several ark shell species are 2–3 times larger than those found in most bilaterians, and are among the largest bilaterian mitochondrial genomes reported to date. These results highlight the need of detailed phylogenetic study to explore evolutionary relationships within Arcoida so that the evolution of mitochondrial genome size can be understood. To this end, we sequenced 17 mitochondrial genomes and compared them with publicly available data, including those from other lineages of Arcoida with emphasis on the subclade Arcoidea species. Our phylogenetic analyses indicate that Noetiidae, Cucullaeidae and Glycymerididae are nested within a polyphyletic Arcidae. Moreover, we find multiple independent expansions and potential contractions of mitochondrial genome size, suggesting that the large mitochondrial genome is not a shared ancestral feature in Arcoida. We also examined tandem repeats and inverted repeats in non-coding regions and investigated the presence of such repeats with relation to genome size variation. Our results suggest that tandem repeats might facilitate intraspecific mitochondrial genome size variation, and that inverted repeats, which could be derived from transposons, might be responsible for mitochondrial genome expansions and contractions. We show that mitochondrial genome size in Arcoida is more dynamic than previously understood and provide insights into evolution of mitochondrial genome size variation in metazoans.
中文翻译:

线粒体基因组学揭示了Arcoida(软体动物,Bivalvia)与线粒体基因组大小的多个独立扩展和收缩的系统发生关系。
Arcoida是大约570种血蛤的一种,是双壳软体动物在生态和经济上很重要的血统。当前对类弧的分类主要基于形态学,这表明广泛的同质性。尽管最近有两项研究采用多位点分析并广泛采样了Arcoida,但主要谱系之间的进化关系仍然存在争议。有趣的是,几种方舟壳物种的线粒体基因组比大多数双语生物中的线粒体基因组大2–3倍,并且是迄今为止报道的最大的双语生物线粒体基因组之一。这些结果强调需要进行详细的系统发育研究,以探索Arcoida内的进化关系,以便可以了解线粒体基因组大小的进化。为此,我们对17个线粒体基因组进行了测序,并将其与公开数据进行了比较,包括来自Arcoida其他谱系的数据,重点是亚科Arcoidea。我们的系统发育分析表明,Noetiidae,Cucullaeidae和Glycymerididae嵌套在多系统Arcidae中。此外,我们发现线粒体基因组大小有多个独立的扩展和潜在的收缩,这表明大的线粒体基因组不是Arcoida中的共同祖先特征。我们还检查了非编码区的串联重复和反向重复,并研究了与基因组大小变化有关的此类重复的存在。我们的结果表明,串联重复序列可能会促进种内线粒体基因组大小的变化,而反向重复序列可能源自转座子,可能负责线粒体基因组的扩增和收缩。我们显示Arcoida中的线粒体基因组大小比以前理解的更动态,并提供对后生动物线粒体基因组大小变异进化的见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号