Molecular Phylogenetics and Evolution ( IF 3.6 ) Pub Date : 2020-05-28 , DOI: 10.1016/j.ympev.2020.106862 W Tyler McCraney 1 , Christine E Thacker 2 , Michael E Alfaro 1
|
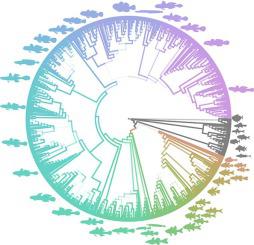
Gobies, sleepers, and cardinalfishes represent major clades of a species rich radiation of small bodied, ecologically diverse percomorphs (Gobiaria). Molecular phylogenetics has been crucial to resolving broad relationships of sleepers and gobies (Gobioidei), but the phylogenetic placements of cardinalfishes and nurseryfishes, as reciprocal or sequential sister clades to Gobioidei, are uncertain. In order to evaluate relationships among and within families we used a phylogenetic data mining approach to generate densely sampled trees inclusive of all higher taxa. We utilized conspecific amino acid homology to improve alignment accuracy, included ambiguously identified taxa to increase taxon sampling density, and resampled individual gene alignments to filter rogue sequences before concatenation. This approach yielded the most comprehensive tree yet of Gobiaria, inferred from a sparse (17 percent-complete) supermatrix of one ribosomal and 22 protein coding loci (18,065 characters), comprised of 50 outgroup and 777 ingroup taxa, representing 32 percent of species and 68 percent of genera. Our analyses confirmed the lineage-based classification of gobies with strong support, identified sleeper clades with unforeseen levels of systematic uncertainty, and quantified competing phylogenetic signals that confound resolution of the root topology. We also discovered that multilocus data completeness was related to maximum likelihood branch support, and verified that the phylogenetic uncertainty of shallow relationships observed within goby lineages could largely be explained by supermatrix sparseness. These results demonstrate the potential and limits of publicly available sequence data for producing densely-sampled phylogenetic trees of exceptionally biodiverse groups.
中文翻译:

超级基质系统发育解析虾虎鱼谱系,并揭示了戈比亚氏菌的不稳定根。
虾虎鱼,睡眠者和红衣主教代表着丰富的物种的主要进化枝,这些物种散布着小巧的,生态多样的percomorphs(戈壁藻)。分子系统发育学对于解决睡眠者和虾虎鱼之间的广泛关系至关重要(Gobioidei),但是,作为枢纽或顺序的姊妹进化枝,对枢纽鱼类和苗鱼的系统发育位置尚不确定。为了评估家庭之间和家庭内部的关系,我们使用了系统发育数据挖掘方法来生成密集采样的树木,其中包括所有较高的分类单元。我们利用同种氨基酸同源性来提高比对准确性,包括模糊鉴定的类群以增加类群采样密度,并重新采样单个基因比对以在级联之前过滤无赖序列。这种方法产生了戈比亚里亚最完整的树,是从一个稀疏的(17%完整)的超级矩阵推断出来的,其中一个矩阵有22个蛋白质编码基因座(18,065个字符),由50个外群和777个内群类群组成,代表了32%的物种和属的68%我们的分析证实了有力支持者对虾虎鱼的基于谱系的分类,确定了具有不可预见水平的系统不确定性的卧铺进化枝,并量化了混淆根拓扑解析度的竞争性系统发生信号。我们还发现多位点数据的完整性与最大似然分支支持有关,并验证了虾虎鱼谱系中观察到的浅层关系的系统发生不确定性很大程度上可以由超级矩阵稀疏性来解释。











































 京公网安备 11010802027423号
京公网安备 11010802027423号