当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Alchemical free energy calculations via metadynamics: Application to the theophylline‐RNA aptamer complex
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2020-05-25 , DOI: 10.1002/jcc.26221 Yoshiaki Tanida 1 , Azuma Matsuura 1
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2020-05-25 , DOI: 10.1002/jcc.26221 Yoshiaki Tanida 1 , Azuma Matsuura 1
Affiliation
|
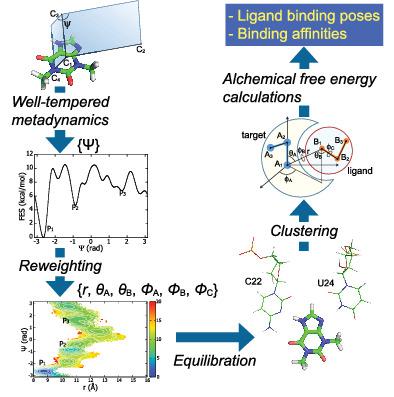
We propose a computational workflow for robust and accurate prediction of both binding poses and their affinities at early stage in designing drug candidates. Small, rigid ligands with few intramolecular degrees of freedom, for example, fragment‐like molecules, have multiple binding poses, even at a single binding site, and their affinities are often close to each other. We explore various structures of ligand binding to a target through metadynamics using a small number of collective variables, followed by reweighting to obtain the atomic coordinates. After identifying each binding pose by cluster analysis, we perform alchemical free energy calculations on each structure to obtain the overall value. We applied this protocol in computing free energy of binding for the theophylline‐RNA aptamer complex. Of the six (meta)stable structures found, the most favorable binding structure is consistent with the structure obtained by NMR. The overall free energy of binding reproduces the experimental values very well.
中文翻译:

通过元动力学计算炼金术自由能:在茶碱-RNA 适配体复合物中的应用
我们提出了一种计算工作流程,用于在设计候选药物的早期阶段对结合姿势及其亲和力进行稳健和准确的预测。分子内自由度很小的小的刚性配体,例如片段样分子,即使在单个结合位点,也具有多种结合姿势,并且它们的亲和力通常彼此接近。我们使用少量集体变量通过元动力学探索与目标结合的配体的各种结构,然后重新加权以获得原子坐标。通过聚类分析确定每个结合位姿后,我们对每个结构进行炼金术自由能计算以获得整体值。我们应用该协议来计算茶碱-RNA 适配体复合物的结合自由能。在发现的六个(元)稳定结构中,最有利的结合结构与核磁共振得到的结构一致。结合的总自由能很好地再现了实验值。
更新日期:2020-05-25
中文翻译:

通过元动力学计算炼金术自由能:在茶碱-RNA 适配体复合物中的应用
我们提出了一种计算工作流程,用于在设计候选药物的早期阶段对结合姿势及其亲和力进行稳健和准确的预测。分子内自由度很小的小的刚性配体,例如片段样分子,即使在单个结合位点,也具有多种结合姿势,并且它们的亲和力通常彼此接近。我们使用少量集体变量通过元动力学探索与目标结合的配体的各种结构,然后重新加权以获得原子坐标。通过聚类分析确定每个结合位姿后,我们对每个结构进行炼金术自由能计算以获得整体值。我们应用该协议来计算茶碱-RNA 适配体复合物的结合自由能。在发现的六个(元)稳定结构中,最有利的结合结构与核磁共振得到的结构一致。结合的总自由能很好地再现了实验值。











































 京公网安备 11010802027423号
京公网安备 11010802027423号