Journal of Biomedical informatics ( IF 4.0 ) Pub Date : 2020-05-23 , DOI: 10.1016/j.jbi.2020.103451 Yu Zhu 1 , Lishuang Li 1 , Hongbin Lu 1 , Anqiao Zhou 1 , Xueyang Qin 1
|
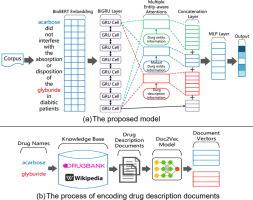
Drug-drug interactions (DDIs) extraction is one of the important tasks in the field of biomedical relation extraction, which plays an important role in the field of pharmacovigilance. Previous neural network based models have achieved good performance in DDIs extraction. However, most of the previous models did not make good use of the information of drug entity names, which can help to judge the relation between drugs. This is mainly because drug names are often very complex, leading to the fact that neural network models cannot understand their semantics directly. To address this issue, we propose a DDIs extraction model using multiple entity-aware attentions with various entity information. We use an output-modified bidirectional transformer (BioBERT) and a bidirectional gated recurrent unit layer (BiGRU) to obtain the vector representation of sentences. The vectors of drug description documents encoded by Doc2Vec are used as drug description information, which is an external knowledge to our model. Then we construct three different kinds of entity-aware attentions to get the sentence representations with entity information weighted, including attentions using the drug description information. The outputs of attention layers are concatenated and fed into a multi-layer perception layer. Finally, we get the result by a softmax classifier. The F-score is used to evaluate our model, which is also adopted by most previous DDIs extraction models. We evaluate our proposed model on the DDIExtraction 2013 corpus, which is the benchmark corpus of this domain, and achieves the state-of-the-art result (80.9% in F-score).
中文翻译:

利用BioBERT从文本中提取药物相互作用,并注意多个实体。
药物相互作用(DDI)的提取是生物医学关系提取领域的重要任务之一,在药物警戒领域起着重要的作用。以前的基于神经网络的模型在DDI提取中已经取得了良好的性能。但是,大多数以前的模型都没有充分利用毒品实体名称的信息,这有助于判断毒品之间的关系。这主要是因为药物名称通常非常复杂,导致神经网络模型无法直接理解其语义。为了解决此问题,我们提出了一种DDI提取模型,该模型使用多个具有各种实体信息的实体感知注意。我们使用输出修改型双向变换器(BioBERT)和双向门控递归单元层(BiGRU)来获取句子的矢量表示。由Doc2Vec编码的药物描述文档的向量用作药物描述信息,这是我们模型的外部知识。然后,我们构造三种不同的实体感知注意,以获取具有实体信息加权的句子表示形式,包括使用药物描述信息进行注意。注意层的输出被串联并馈入多层感知层。最后,我们通过softmax分类器获得结果。F分数用于评估我们的模型,大多数先前的DDI提取模型也采用了该分数。我们在DDIExtraction 2013语料库上评估我们提出的模型,











































 京公网安备 11010802027423号
京公网安备 11010802027423号