当前位置:
X-MOL 学术
›
Ecol. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
In silico and empirical evaluation of twelve metabarcoding primer sets for insectivorous diet analyses.
Ecology and Evolution ( IF 2.3 ) Pub Date : 2020-05-21 , DOI: 10.1002/ece3.6362 Orianne Tournayre 1 , Maxime Leuchtmann 2 , Ondine Filippi-Codaccioni 3, 4 , Marine Trillat 1 , Sylvain Piry 1 , Dominique Pontier 3, 4 , Nathalie Charbonnel 1 , Maxime Galan 1
Ecology and Evolution ( IF 2.3 ) Pub Date : 2020-05-21 , DOI: 10.1002/ece3.6362 Orianne Tournayre 1 , Maxime Leuchtmann 2 , Ondine Filippi-Codaccioni 3, 4 , Marine Trillat 1 , Sylvain Piry 1 , Dominique Pontier 3, 4 , Nathalie Charbonnel 1 , Maxime Galan 1
Affiliation
|
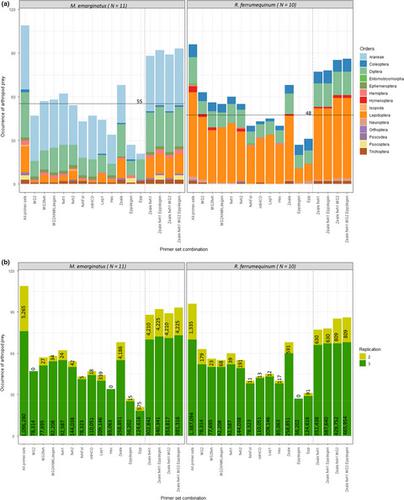
During the most recent decade, environmental DNA metabarcoding approaches have been both developed and improved to minimize the biological and technical biases in these protocols. However, challenges remain, notably those relating to primer design. In the current study, we comprehensively assessed the performance of ten COI and two 16S primer pairs for eDNA metabarcoding, including novel and previously published primers. We used a combined approach of in silico, in vivo‐mock community (33 arthropod taxa from 16 orders), and guano‐based analyses to identify primer sets that would maximize arthropod detection and taxonomic identification, successfully identify the predator (bat) species, and minimize the time and financial costs of the experiment. We focused on two insectivorous bat species that live together in mixed colonies: the greater horseshoe bat (Rhinolophus ferrumequinum ) and Geoffroy's bat (Myotis emarginatus ). We found that primer degeneracy is the main factor that influences arthropod detection in silico and mock community analyses, while amplicon length is critical for the detection of arthropods from degraded DNA samples. Our guano‐based results highlight the importance of detecting and identifying both predator and prey, as guano samples can be contaminated by other insectivorous species. Moreover, we demonstrate that amplifying bat DNA does not reduce the primers' capacity to detect arthropods. We therefore recommend the simultaneous identification of predator and prey. Finally, our results suggest that up to one‐third of prey occurrences may be unreliable and are probably not of primary interest in diet studies, which may decrease the relevance of combining several primer sets instead of using a single efficient one. In conclusion, this study provides a pragmatic framework for eDNA primer selection with respect to scientific and methodological constraints.
中文翻译:

用于食虫饮食分析的十二个元条形码引物组的计算机模拟和实证评估。
在最近十年中,环境 DNA 元条形码方法得到了发展和改进,以最大限度地减少这些方案中的生物学和技术偏差。然而,挑战仍然存在,特别是与引物设计相关的挑战。在当前的研究中,我们全面评估了 10 个 COI 和两个 16S 引物对用于 eDNA 元条形码的性能,包括新颖的和先前发表的引物。我们采用了计算机模拟、体内模拟群落(来自 16 个目的 33 个节肢动物类群)和基于鸟粪的分析相结合的方法来确定引物组,以最大限度地提高节肢动物检测和分类学鉴定,成功识别捕食者(蝙蝠)物种,并最大限度地减少实验的时间和财务成本。我们重点关注两种生活在混合群体中的食虫蝙蝠:大马蹄蝠 ( Rhinolophus ferrumequinum ) 和杰弗罗伊蝙蝠 ( Myotis emarginatus )。我们发现引物简并性是影响计算机和模拟群落分析中节肢动物检测的主要因素,而扩增子长度对于从降解的 DNA 样本中检测节肢动物至关重要。我们基于鸟粪的结果强调了检测和识别捕食者和猎物的重要性,因为鸟粪样本可能被其他食虫物种污染。此外,我们证明扩增蝙蝠 DNA 不会降低引物检测节肢动物的能力。因此,我们建议同时识别捕食者和猎物。最后,我们的结果表明,多达三分之一的猎物出现可能不可靠,并且可能不是饮食研究的主要兴趣,这可能会降低组合多个引物组而不是使用单个有效引物组的相关性。 总之,本研究为 eDNA 引物选择提供了一个在科学和方法学限制方面的务实框架。
更新日期:2020-07-25
中文翻译:

用于食虫饮食分析的十二个元条形码引物组的计算机模拟和实证评估。
在最近十年中,环境 DNA 元条形码方法得到了发展和改进,以最大限度地减少这些方案中的生物学和技术偏差。然而,挑战仍然存在,特别是与引物设计相关的挑战。在当前的研究中,我们全面评估了 10 个 COI 和两个 16S 引物对用于 eDNA 元条形码的性能,包括新颖的和先前发表的引物。我们采用了计算机模拟、体内模拟群落(来自 16 个目的 33 个节肢动物类群)和基于鸟粪的分析相结合的方法来确定引物组,以最大限度地提高节肢动物检测和分类学鉴定,成功识别捕食者(蝙蝠)物种,并最大限度地减少实验的时间和财务成本。我们重点关注两种生活在混合群体中的食虫蝙蝠:大马蹄蝠 ( Rhinolophus ferrumequinum ) 和杰弗罗伊蝙蝠 ( Myotis emarginatus )。我们发现引物简并性是影响计算机和模拟群落分析中节肢动物检测的主要因素,而扩增子长度对于从降解的 DNA 样本中检测节肢动物至关重要。我们基于鸟粪的结果强调了检测和识别捕食者和猎物的重要性,因为鸟粪样本可能被其他食虫物种污染。此外,我们证明扩增蝙蝠 DNA 不会降低引物检测节肢动物的能力。因此,我们建议同时识别捕食者和猎物。最后,我们的结果表明,多达三分之一的猎物出现可能不可靠,并且可能不是饮食研究的主要兴趣,这可能会降低组合多个引物组而不是使用单个有效引物组的相关性。 总之,本研究为 eDNA 引物选择提供了一个在科学和方法学限制方面的务实框架。










































 京公网安备 11010802027423号
京公网安备 11010802027423号