Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
CRISPR‐Net: A Recurrent Convolutional Network Quantifies CRISPR Off‐Target Activities with Mismatches and Indels
Advanced Science ( IF 15.1 ) Pub Date : 2020-05-20 , DOI: 10.1002/advs.201903562 Jiecong Lin 1 , Zhaolei Zhang 2, 3, 4 , Shixiong Zhang 1 , Junyi Chen 1 , Ka‐Chun Wong 1
Advanced Science ( IF 15.1 ) Pub Date : 2020-05-20 , DOI: 10.1002/advs.201903562 Jiecong Lin 1 , Zhaolei Zhang 2, 3, 4 , Shixiong Zhang 1 , Junyi Chen 1 , Ka‐Chun Wong 1
Affiliation
|
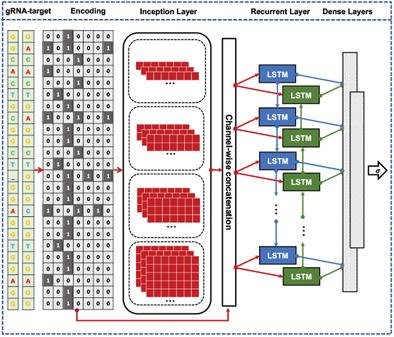
The off‐target effects induced by guide RNAs in the CRISPR/Cas9 gene‐editing system have raised substantial concerns in recent years. Many in silico predictive models have been developed for predicting the off‐target activities; however, few are capable of predicting the off‐target activities with insertions or deletions between guide RNA and target DNA sequence pair. In order to fill this gap, a recurrent convolutional network named CRISPR‐Net is developed for scoring the gRNA‐target pairs with mismatches and indels; and a machine‐learning based model named CRISPR‐Net‐Aggregate is also developed for aggregating the scores as the consensus off‐target score for each potential guide RNA. It is demonstrated that CRISPR‐Net achieves competitive performance on CIRCLE‐Seq and GUIDE‐seq datasets with indels and mismatches, outperforming the state‐of‐the‐art off‐target prediction methods on two independent mismatch‐only datasets. The CRISPR‐Net‐Aggregate also surpasses a competing method on the aggregation task. Moreover, a two‐stage sensitivity analysis is introduced to visualize the CRISPR‐Net prediction on the gRNA‐target pair of interest, demonstrating how implicit knowledge encoded in CRISPR‐Net contributes to the accurate off‐target activity quantification. Finally, the source code is made available at the Code Ocean repository (https://codeocean.com/capsule/9553651/tree/v1).
中文翻译:

CRISPR-Net:循环卷积网络可通过错配和插入缺失量化CRISPR脱靶活动
近年来,由CRISPR / Cas9基因编辑系统中的引导RNA引起的脱靶效应引起了人们的广泛关注。已经开发出许多计算机模拟预测模型来预测脱靶活动。但是,很少有人能够通过指导RNA和靶DNA序列对之间的插入或缺失来预测脱靶活动。为了填补这一空白,开发了一个名为CRISPR-Net的循环卷积网络,用于对具有错配和插入缺失的gRNA-靶标对进行评分。并且还开发了一种基于机器学习的名为CRISPR-Net-Aggregate的模型,用于将分数汇总为每个潜在指导RNA的共识脱靶分数。事实证明,CRISPR-Net在CIRCLE-Seq和GUIDE-seq数据集上具有插入缺失和错配的竞争优势,在两个独立的仅失配数据集上,它们的表现优于最新的脱靶预测方法。CRISPR-Net-Aggregate在聚合任务上也超越了竞争方法。此外,引入了两阶段敏感性分析以可视化对感兴趣的gRNA-靶标对的CRISPR-Net预测,证明CRISPR-Net中编码的隐式知识如何有助于精确的脱靶活性定量。最后,可以在Code Ocean存储库(https://codeocean.com/capsule/9553651/tree/v1)中获得源代码。引入了两阶段敏感性分析以可视化对感兴趣的gRNA-靶标对的CRISPR-Net预测,证明CRISPR-Net中编码的隐式知识如何有助于精确的脱靶活性定量。最后,可在Code Ocean存储库(https://codeocean.com/capsule/9553651/tree/v1)中获得源代码。引入了两阶段敏感性分析以可视化对感兴趣的gRNA-靶标对的CRISPR-Net预测,证明CRISPR-Net中编码的隐式知识如何有助于精确的脱靶活性定量。最后,可在Code Ocean存储库(https://codeocean.com/capsule/9553651/tree/v1)中获得源代码。
更新日期:2020-07-08
中文翻译:

CRISPR-Net:循环卷积网络可通过错配和插入缺失量化CRISPR脱靶活动
近年来,由CRISPR / Cas9基因编辑系统中的引导RNA引起的脱靶效应引起了人们的广泛关注。已经开发出许多计算机模拟预测模型来预测脱靶活动。但是,很少有人能够通过指导RNA和靶DNA序列对之间的插入或缺失来预测脱靶活动。为了填补这一空白,开发了一个名为CRISPR-Net的循环卷积网络,用于对具有错配和插入缺失的gRNA-靶标对进行评分。并且还开发了一种基于机器学习的名为CRISPR-Net-Aggregate的模型,用于将分数汇总为每个潜在指导RNA的共识脱靶分数。事实证明,CRISPR-Net在CIRCLE-Seq和GUIDE-seq数据集上具有插入缺失和错配的竞争优势,在两个独立的仅失配数据集上,它们的表现优于最新的脱靶预测方法。CRISPR-Net-Aggregate在聚合任务上也超越了竞争方法。此外,引入了两阶段敏感性分析以可视化对感兴趣的gRNA-靶标对的CRISPR-Net预测,证明CRISPR-Net中编码的隐式知识如何有助于精确的脱靶活性定量。最后,可以在Code Ocean存储库(https://codeocean.com/capsule/9553651/tree/v1)中获得源代码。引入了两阶段敏感性分析以可视化对感兴趣的gRNA-靶标对的CRISPR-Net预测,证明CRISPR-Net中编码的隐式知识如何有助于精确的脱靶活性定量。最后,可在Code Ocean存储库(https://codeocean.com/capsule/9553651/tree/v1)中获得源代码。引入了两阶段敏感性分析以可视化对感兴趣的gRNA-靶标对的CRISPR-Net预测,证明CRISPR-Net中编码的隐式知识如何有助于精确的脱靶活性定量。最后,可在Code Ocean存储库(https://codeocean.com/capsule/9553651/tree/v1)中获得源代码。



























 京公网安备 11010802027423号
京公网安备 11010802027423号