Journal of Allergy and Clinical Immunology ( IF 11.4 ) Pub Date : 2020-05-19 , DOI: 10.1016/j.jaci.2020.04.051 Maria E Ketelaar 1 , Michael A Portelli 2 , F Nicole Dijk 3 , Nick Shrine 4 , Alen Faiz 5 , Cornelis J Vermeulen 5 , Cheng J Xu 6 , Jenny Hankinson 7 , Sangita Bhaker 2 , Amanda P Henry 2 , Charlote K Billington 2 , Dominick E Shaw 2 , Simon R Johnson 2 , Andrew V Benest 8 , Vincent Pang 8 , David O Bates 8 , Z E K Pogson 9 , Andrew Fogarty 10 , Tricia M McKeever 10 , Amisha Singapuri 11 , Liam G Heaney 12 , Adel H Mansur 13 , Rekha Chaudhuri 14 , Neil C Thomson 14 , John W Holloway 15 , Gabrielle A Lockett 15 , Peter H Howarth 15 , Robert Niven 7 , Angela Simpson 7 , Martin D Tobin 4 , Ian P Hall 2 , Louise V Wain 4 , John D Blakey 16 , Christopher E Brightling 17 , Ma'en Obeidat 18 , Don D Sin 19 , David C Nickle 20 , Yohan Bossé 21 , Judith M Vonk 22 , Maarten van den Berge 23 , Gerard H Koppelman 3 , Ian Sayers 2 , Martijn C Nawijn 24
|
Background
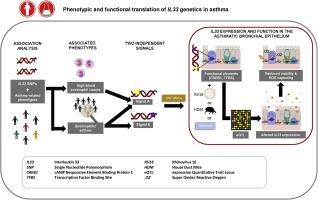
Asthma is a complex disease with multiple phenotypes that may differ in disease pathobiology and treatment response. IL33 single nucleotide polymorphisms (SNPs) have been reproducibly associated with asthma. IL33 levels are elevated in sputum and bronchial biopsies of patients with asthma. The functional consequences of IL33 asthma SNPs remain unknown.
Objective
This study sought to determine whether IL33 SNPs associate with asthma-related phenotypes and with IL33 expression in lung or bronchial epithelium. This study investigated the effect of increased IL33 expression on human bronchial epithelial cell (HBEC) function.
Methods
Association between IL33 SNPs (Chr9: 5,815,786-6,657,983) and asthma phenotypes (Lifelines/DAG [Dutch Asthma GWAS]/GASP [Genetics of Asthma Severity & Phenotypes] cohorts) and between SNPs and expression (lung tissue, bronchial brushes, HBECs) was done using regression modeling. Lentiviral overexpression was used to study IL33 effects on HBECs.
Results
We found that 161 SNPs spanning the IL33 region associated with 1 or more asthma phenotypes after correction for multiple testing. We report a main independent signal tagged by rs992969 associating with blood eosinophil levels, asthma, and eosinophilic asthma. A second, independent signal tagged by rs4008366 presented modest association with eosinophilic asthma. Neither signal associated with FEV1, FEV1/forced vital capacity, atopy, and age of asthma onset. The 2 IL33 signals are expression quantitative loci in bronchial brushes and cultured HBECs, but not in lung tissue. IL33 overexpression in vitro resulted in reduced viability and reactive oxygen species–capturing of HBECs, without influencing epithelial cell count, metabolic activity, or barrier function.
Conclusions
We identify IL33 as an epithelial susceptibility gene for eosinophilia and asthma, provide mechanistic insight, and implicate targeting of the IL33 pathway specifically in eosinophilic asthma.
中文翻译:

IL33遗传学在哮喘中的表型和功能翻译。
背景
哮喘是一种复杂的疾病,具有多种表型,在疾病病理生物学和治疗反应方面可能不同。IL33单核苷酸多态性 (SNP) 与哮喘具有可重复性相关。哮喘患者的痰液和支气管活检中 IL33 水平升高。IL33哮喘 SNP的功能后果仍然未知。
客观的
本研究旨在确定IL33 SNP 是否与哮喘相关表型以及肺或支气管上皮细胞中的IL33表达相关。本研究调查了增加的IL33表达对人支气管上皮细胞 (HBEC) 功能的影响。
方法
IL33 SNP(Chr9:5,815,786-6,657,983)与哮喘表型(Lifelines/DAG [荷兰哮喘 GWAS]/GASP [哮喘严重程度和表型的遗传学] 队列)之间以及 SNP 与表达(肺组织、支气管刷、HBEC)之间的关联是使用回归模型完成。慢病毒过表达用于研究IL33 对HBEC 的影响。
结果
我们发现,经过多次测试校正后,跨越IL33区域的 161 个 SNP 与 1 种或多种哮喘表型相关。我们报告了一个由 rs992969 标记的主要独立信号,与血液嗜酸性粒细胞水平、哮喘和嗜酸性粒细胞性哮喘相关。由 rs4008366 标记的第二个独立信号与嗜酸性粒细胞性哮喘有适度关联。信号均与 FEV 1、FEV 1 /用力肺活量、特应性和哮喘发作年龄无关。2 个 IL33 信号是支气管刷和培养的HBEC中的表达定量基因座,但在肺组织中没有。IL33体外过表达导致 HBEC 的活力和活性氧物种捕获降低,而不影响上皮细胞计数、代谢活性或屏障功能。
结论
我们将IL33鉴定为嗜酸性粒细胞增多症和哮喘的上皮易感基因,提供机制见解,并暗示 IL33 通路的靶向特异性在嗜酸性粒细胞性哮喘中。











































 京公网安备 11010802027423号
京公网安备 11010802027423号