当前位置:
X-MOL 学术
›
J. Zool. Syst. Evol. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Species delimitation through DNA barcoding of freshwater leeches of the Glossiphonia genus (Hirudinea: Glossiphoniidae) from Eastern Siberia, Russia
Journal of Zoological Systematics and Evolutionary Research ( IF 2.0 ) Pub Date : 2020-05-17 , DOI: 10.1111/jzs.12385 Irina Kaygorodova 1 , Nadezhda Bolbat 1, 2 , Alexander Bolbat 1
Journal of Zoological Systematics and Evolutionary Research ( IF 2.0 ) Pub Date : 2020-05-17 , DOI: 10.1111/jzs.12385 Irina Kaygorodova 1 , Nadezhda Bolbat 1, 2 , Alexander Bolbat 1
Affiliation
|
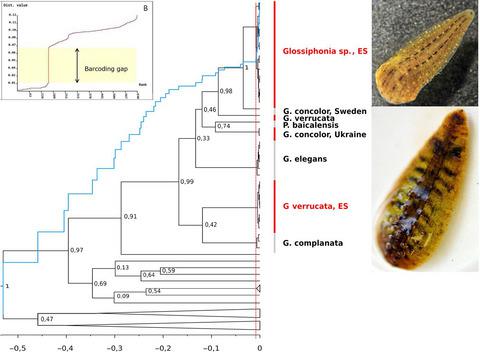
The study of biodiversity is a priority task of biological science. The structural unit of biodiversity is a species that has a clear identification in a taxonomic system. Morphological features are traditionally the main criteria for species discrimination in zoological studies. However, the presence of inter‐ and intraspecific polymorphism and phenotypic plasticity makes it difficult to identify species in many groups of invertebrates. To solve this problem, in this research, we analyzed morphological and genetic data in combination to delimit species among the Eastern Siberia Glossiphonia leeches using different approaches. Morphology analysis revealed phenetically distinct groups, suggesting the existence of at least two species in the region, G. verrucata, a rare Palaearctic species, and a potentially new species Glossiphonia sp. Moreover, sequence‐based species delimitation methods congruently supported eight distinct species groups (including two Siberian species) within the available molecular dataset of the Glossiphonia world fauna, using phylogenetic (ML and BI), coalescent (ABGD and GMYC) methods, and pairwise analysis of sequences. The detected p‐distances (modal value of 0.11) between these 8 groups and the level of genetic polymorphism (max. 0.0041) within groups indicate that the groups are 8 independent species according to the DNA barcoding. Our results once again proved the usefulness of molecular systematics. At the same time, we detected several inaccuracies in the leech species identification, as well as many ambiguous sites in sequences uploaded on GenBank, which affects the analysis and impedes progress of DNA barcoding technology.
中文翻译:

通过对俄罗斯西伯利亚东部的Glossiphonia属(Hirudinea:Glossiphoniidae)的淡水水DNA进行DNA条形码划分物种。
生物多样性的研究是生物科学的优先任务。生物多样性的结构单位是在分类系统中具有明确标识的物种。传统上,形态特征是动物学研究中判别物种的主要标准。然而,种间和种内多态性和表型可塑性的存在使得很难在许多无脊椎动物组中鉴定物种。为了解决这个问题,在本研究中,我们结合不同的方法分析了形态和遗传数据,以界定东西伯利亚Glossiphonia水中的物种。形态学分析揭示了在形态上不同的群体,这表明该地区存在至少两种物种,即疣状拟南芥,稀有的古北种,还有潜在的新种Glossiphonia sp。此外,基于序列的物种划界方法可在Glossiphonia的可用分子数据集中全力支持八个不同的物种组(包括两个西伯利亚物种)世界动物区系,使用系统发育(ML和BI),合并(ABGD和GMYC)方法以及成对序列分析。在这8个组之间检测到的p距离(模式值0.11)和组内的遗传多态性水平(最大0.0041)表明,根据DNA条形码,这些组是8个独立的物种。我们的结果再次证明了分子系统学的有用性。同时,我们发现了水ech物种鉴定中的一些错误,以及在GenBank上载的序列中的许多歧义位点,这影响了分析并阻碍了DNA条形码技术的进步。
更新日期:2020-05-17
中文翻译:

通过对俄罗斯西伯利亚东部的Glossiphonia属(Hirudinea:Glossiphoniidae)的淡水水DNA进行DNA条形码划分物种。
生物多样性的研究是生物科学的优先任务。生物多样性的结构单位是在分类系统中具有明确标识的物种。传统上,形态特征是动物学研究中判别物种的主要标准。然而,种间和种内多态性和表型可塑性的存在使得很难在许多无脊椎动物组中鉴定物种。为了解决这个问题,在本研究中,我们结合不同的方法分析了形态和遗传数据,以界定东西伯利亚Glossiphonia水中的物种。形态学分析揭示了在形态上不同的群体,这表明该地区存在至少两种物种,即疣状拟南芥,稀有的古北种,还有潜在的新种Glossiphonia sp。此外,基于序列的物种划界方法可在Glossiphonia的可用分子数据集中全力支持八个不同的物种组(包括两个西伯利亚物种)世界动物区系,使用系统发育(ML和BI),合并(ABGD和GMYC)方法以及成对序列分析。在这8个组之间检测到的p距离(模式值0.11)和组内的遗传多态性水平(最大0.0041)表明,根据DNA条形码,这些组是8个独立的物种。我们的结果再次证明了分子系统学的有用性。同时,我们发现了水ech物种鉴定中的一些错误,以及在GenBank上载的序列中的许多歧义位点,这影响了分析并阻碍了DNA条形码技术的进步。











































 京公网安备 11010802027423号
京公网安备 11010802027423号