当前位置:
X-MOL 学术
›
Chem. Res. Toxicol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Ensemble Models Based on QuBiLS-MAS Features and Shallow Learning for the Prediction of Drug-Induced Liver Toxicity: Improving Deep Learning and Traditional Approaches.
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2020-05-14 , DOI: 10.1021/acs.chemrestox.0c00030 Jose R Mora 1, 2 , Yovani Marrero-Ponce 2, 3 , César R García-Jacas 4 , Amileth Suarez Causado 5
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2020-05-14 , DOI: 10.1021/acs.chemrestox.0c00030 Jose R Mora 1, 2 , Yovani Marrero-Ponce 2, 3 , César R García-Jacas 4 , Amileth Suarez Causado 5
Affiliation
|
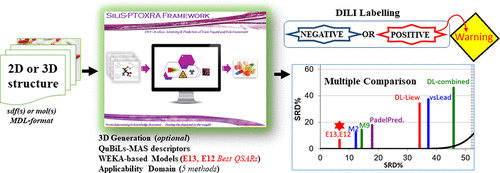
Drug-induced liver injury (DILI) is a key safety issue in the drug discovery pipeline and a regulatory concern. Thus, many in silico tools have been proposed to improve the hepatotoxicity prediction of organic-type chemicals. Here, classifiers for the prediction of DILI were developed by using QuBiLS-MAS 0–2.5D molecular descriptors and shallow machine learning techniques, on a training set composed of 1075 molecules. The best ensemble model build, E13, was obtained with good statistical parameters for the learning series, namely, the following: accuracy = 0.840, sensibility = 0.890, specificity = 0.761, Matthew’s correlation coefficient = 0.660, and area under the ROC curve = 0.904. The model was also satisfactorily evaluated with Y-scrambling test, and repeated k-fold cross-validation and repeated k-holdout validation. In addition, an exhaustive external validation was also carried out by using two test sets and five external test sets, with an average accuracy value equal to 0.854 (±0.062) and a coverage equal to 98.4% according to its applicability domain. A statistical comparison of the performance of the E13 model, with regard to results and tools (e.g., Padel DDPredictor Software, Deep Learning DILIserver, and Vslead) reported in the literature, was also performed. In general, E13 presented the best global performance in all experiments. The sum of the ranking differences procedure provided a very similar grouping pattern to that of the M-ANOVA statistical analysis, where E13 was identified as the best model for DILI predictions. A noncommercial and fully cross-platform software for the DILI prediction was also developed, which is freely available at http://tomocomd.com/apps/ptoxra. This software was used for the screening of seven data sets, containing natural products, leads, toxic materials, and FDA approved drugs, to assess the usefulness of the QSAR models in the DILI labeling of organic substances; it was found that 50–92% of the evaluated molecules are positive-DILI compounds. All in all, it can be stated that the E13 model is a relevant method for the prediction of DILI risk in humans, as it shows the best results among all of the methods analyzed.
中文翻译:

基于QuBiLS-MAS特征和浅层学习的集成模型,用于预测药物引起的肝毒性:改进深度学习和传统方法。
药物诱发的肝损伤(DILI)是药物开发过程中的关键安全问题,也是监管机构关注的问题。因此,已经提出了许多计算机模拟工具来改善有机型化学药品的肝毒性预测。在这里,在由1075个分子组成的训练集上,使用QuBiLS-MAS 0-2.5D分子描述符和浅层机器学习技术开发了用于预测DILI的分类器。对于学习系列,获得具有最佳统计参数的最佳集成模型构建E13,即:精度= 0.840,灵敏度= 0.890,特异性= 0.761,马修相关系数= 0.660,ROC曲线下面积= 0.904 。使用Y对模型也进行了令人满意的评估-加扰测试,以及重复的k-倍交叉验证和重复的k-保持验证。另外,还通过使用两个测试集和五个外部测试集进行了详尽的外部验证,根据其适用范围,平均准确度值等于0.854(±0.062),覆盖率等于98.4%。还对文献中报告的结果和工具(例如Padel DDPredictor软件,深度学习DILIserver和Vslead)进行了E13模型性能的统计比较。一般来说,E13展示了所有实验中最佳的整体性能。排名差异程序的总和提供了与M-ANOVA统计分析非常相似的分组模式,其中E13被确定为DILI预测的最佳模型。还开发了用于DILI预测的非商业性且完全跨平台的软件,该软件可从http://tomocomd.com/apps/ptoxra免费获得。该软件用于筛选七个数据集,包括天然产物,铅,有毒物质和FDA批准的药物,以评估QSAR模型在有机物质DILI标记中的有用性;发现被评估分子中有50–92%是正DILI化合物。总而言之,可以说E13 该模型是预测人类DILI风险的一种相关方法,因为它在所有分析方法中均显示出最佳结果。
更新日期:2020-07-20
中文翻译:

基于QuBiLS-MAS特征和浅层学习的集成模型,用于预测药物引起的肝毒性:改进深度学习和传统方法。
药物诱发的肝损伤(DILI)是药物开发过程中的关键安全问题,也是监管机构关注的问题。因此,已经提出了许多计算机模拟工具来改善有机型化学药品的肝毒性预测。在这里,在由1075个分子组成的训练集上,使用QuBiLS-MAS 0-2.5D分子描述符和浅层机器学习技术开发了用于预测DILI的分类器。对于学习系列,获得具有最佳统计参数的最佳集成模型构建E13,即:精度= 0.840,灵敏度= 0.890,特异性= 0.761,马修相关系数= 0.660,ROC曲线下面积= 0.904 。使用Y对模型也进行了令人满意的评估-加扰测试,以及重复的k-倍交叉验证和重复的k-保持验证。另外,还通过使用两个测试集和五个外部测试集进行了详尽的外部验证,根据其适用范围,平均准确度值等于0.854(±0.062),覆盖率等于98.4%。还对文献中报告的结果和工具(例如Padel DDPredictor软件,深度学习DILIserver和Vslead)进行了E13模型性能的统计比较。一般来说,E13展示了所有实验中最佳的整体性能。排名差异程序的总和提供了与M-ANOVA统计分析非常相似的分组模式,其中E13被确定为DILI预测的最佳模型。还开发了用于DILI预测的非商业性且完全跨平台的软件,该软件可从http://tomocomd.com/apps/ptoxra免费获得。该软件用于筛选七个数据集,包括天然产物,铅,有毒物质和FDA批准的药物,以评估QSAR模型在有机物质DILI标记中的有用性;发现被评估分子中有50–92%是正DILI化合物。总而言之,可以说E13 该模型是预测人类DILI风险的一种相关方法,因为它在所有分析方法中均显示出最佳结果。











































 京公网安备 11010802027423号
京公网安备 11010802027423号