当前位置:
X-MOL 学术
›
Mol. Inform.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Privileged Scaffold Analysis of Natural Products with Deep Learning-based Indication Prediction Model.
Molecular Informatics ( IF 2.8 ) Pub Date : 2020-05-14 , DOI: 10.1002/minf.202000057 Junyong Lai 1 , Jianxing Hu 1 , Yanxing Wang 1 , Xin Zhou 1 , Yibo Li 2 , Liangren Zhang 1 , Zhenming Liu 1
Molecular Informatics ( IF 2.8 ) Pub Date : 2020-05-14 , DOI: 10.1002/minf.202000057 Junyong Lai 1 , Jianxing Hu 1 , Yanxing Wang 1 , Xin Zhou 1 , Yibo Li 2 , Liangren Zhang 1 , Zhenming Liu 1
Affiliation
|
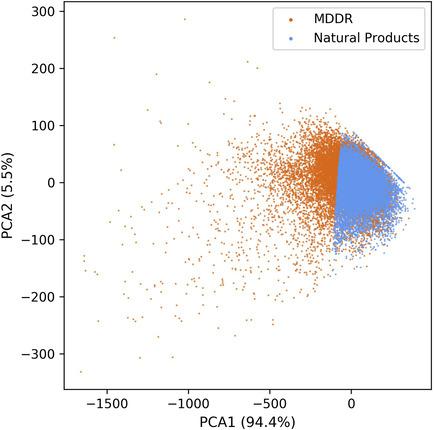
Natural products play a vital role in the drug discovery and development process as an important source of reliable and novel lead structures. But the existing criteria for drug leads were usually developed for synthetic compounds and cannot be directly applied to identify lead scaffolds from natural products. To solve this problem, we propose a method to predict indications and identify privileged scaffolds of natural products for drug design. A deep learning model was built to predict indications for natural products. Entropy‐based information metrics were used to identify the privileged scaffolds for each indication and a Privileged Scaffold Dataset (PSD) of natural products was constructed. The PSD could serve as a novel source of lead compounds and circumvent existing drug patents. This method could be generalized by replacing the training set, the prediction algorithm, and the compound set, to obtain more personalized‐PSDs.
中文翻译:

使用基于深度学习的适应症预测模型对天然产物进行特权支架分析。
天然产物作为可靠和新型先导结构的重要来源,在药物发现和开发过程中发挥着至关重要的作用。但是现有的药物先导标准通常是针对合成化合物制定的,不能直接应用于从天然产物中识别铅支架。为了解决这个问题,我们提出了一种方法来预测适应症并识别用于药物设计的天然产物的特权支架。建立了一个深度学习模型来预测天然产品的适应症。使用基于熵的信息度量来识别每个适应症的特权支架,并构建了天然产物的特权支架数据集 (PSD)。PSD 可以作为先导化合物的新来源并规避现有的药物专利。
更新日期:2020-05-14
中文翻译:

使用基于深度学习的适应症预测模型对天然产物进行特权支架分析。
天然产物作为可靠和新型先导结构的重要来源,在药物发现和开发过程中发挥着至关重要的作用。但是现有的药物先导标准通常是针对合成化合物制定的,不能直接应用于从天然产物中识别铅支架。为了解决这个问题,我们提出了一种方法来预测适应症并识别用于药物设计的天然产物的特权支架。建立了一个深度学习模型来预测天然产品的适应症。使用基于熵的信息度量来识别每个适应症的特权支架,并构建了天然产物的特权支架数据集 (PSD)。PSD 可以作为先导化合物的新来源并规避现有的药物专利。











































 京公网安备 11010802027423号
京公网安备 11010802027423号