Structure ( IF 4.4 ) Pub Date : 2020-05-14 , DOI: 10.1016/j.str.2020.04.018 Michael J Robertson 1 , Gydo C P van Zundert 2 , Kenneth Borrelli 2 , Georgios Skiniotis 3
|
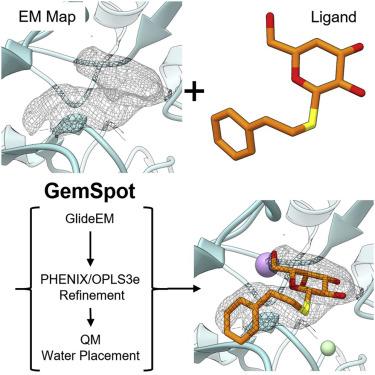
Producing an accurate atomic model of biomolecule-ligand interactions from maps generated by cryoelectron microscopy (cryo-EM) often presents challenges inherent to the methodology and the dynamic nature of ligand binding. Here, we present GemSpot, an automated pipeline of computational chemistry methods that take into account EM map potentials, quantum mechanics energy calculations, and water molecule site prediction to generate candidate poses and provide a measure of the degree of confidence. The pipeline is validated through several published cryo-EM structures of complexes in different resolution ranges and various types of ligands. In all cases, at least one identified pose produced both excellent interactions with the target and agreement with the map. GemSpot will be valuable for the robust identification of ligand poses and drug discovery efforts through cryo-EM.
中文翻译:

GemSpot:用于将配体可靠地建模为Cryo-EM Maps的管道。
从冷冻电子显微镜(cryo-EM)生成的图谱中产生生物分子-配体相互作用的精确原子模型通常会带来配体结合方法学和动力学性质固有的挑战。在这里,我们介绍GemSpot,这是一种计算化学方法的自动化管道,其中考虑了EM映射势,量子力学能量计算和水分子位点预测,以生成候选姿态并提供置信度的度量。通过在不同分辨率范围和各种配体类型下公布的几种复杂的冷冻EM结构,对管道进行了验证。在所有情况下,至少一个识别出的姿势既能与目标互动,又能与地图达成一致。











































 京公网安备 11010802027423号
京公网安备 11010802027423号