Journal of Inorganic Biochemistry ( IF 3.8 ) Pub Date : 2020-05-11 , DOI: 10.1016/j.jinorgbio.2020.111091 John J Kozak 1 , Harry B Gray 2 , Roberto A Garza-López 3
|
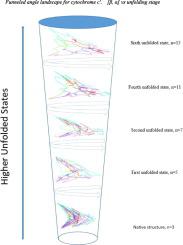
We use crystallographic data for four helical iron proteins (cytochrome c-b562, cytochrome c′, sperm whale myoglobin, human cytoglobin) to calculate radial and angular signatures as each unfolds from the native state stepwise though four unfolded states. From these data we construct an angle phase diagram to display the evolution of each protein from its native state; and, in turn, the phase diagram is used to construct a funneled angle landscape for comparison with the topography of its folding energy landscape. We quantify the departure of individual helical and turning regions from the areal, angular profile of corresponding regions of the native state. This procedure allows us to identify the similarities and differences among individual helical and turning regions in the early stages of unfolding of the four helical heme proteins.
中文翻译:

螺旋形蛋白质的漏斗形景观。
我们使用四种螺旋铁蛋白(细胞色素c -b 562,细胞色素c``,抹香鲸的肌红蛋白,人类细胞红蛋白)来计算径向和角度特征,因为每个特征都从天然状态逐步展开到四个展开状态。根据这些数据,我们构建了一个角度相图,以显示每种蛋白质从其天然状态的进化;然后,将相图用于构建漏斗形的角度景观,以与其折叠能量景观的地形进行比较。我们从自然状态的相应区域的面积,角度轮廓量化了单个螺旋和转向区域的偏离。该程序使我们能够在四个螺旋血红素蛋白展开的早期阶段识别各个螺旋区和转向区之间的相似性和差异。











































 京公网安备 11010802027423号
京公网安备 11010802027423号