当前位置:
X-MOL 学术
›
Acta Cryst. D
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Structure-function study of AKR4C14, an aldo-keto reductase from Thai jasmine rice (Oryza sativa L. ssp. indica cv. KDML105).
Acta Crystallographica Section D ( IF 2.2 ) Pub Date : 2020-05-01 , DOI: 10.1107/s2059798320004313 Chomphunuch Songsiriritthigul , Rawint Narawongsanont , Chonticha Tantitadapitak , Hong-Hsiang Guan , Chun-Jung Chen
Acta Crystallographica Section D ( IF 2.2 ) Pub Date : 2020-05-01 , DOI: 10.1107/s2059798320004313 Chomphunuch Songsiriritthigul , Rawint Narawongsanont , Chonticha Tantitadapitak , Hong-Hsiang Guan , Chun-Jung Chen
|
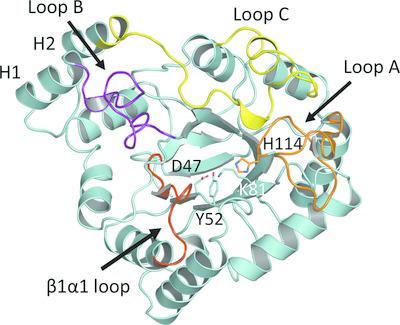
Aldo‐keto reductases (AKRs) are NADPH/NADP+‐dependent oxidoreductase enzymes that metabolize an aldehyde/ketone to the corresponding alcohol. AKR4C14 from rice exhibits a much higher efficiency in metabolizing malondialdehyde (MDA) than do the Arabidopsis enzymes AKR4C8 and AKR4C9, despite sharing greater than 60% amino‐acid sequence identity. This study confirms the role of rice AKR4C14 in the detoxification of methylglyoxal and MDA, and demonstrates that the endogenous contents of both aldehydes in transgenic Arabidopsis ectopically expressing AKR4C14 are significantly lower than their levels in the wild type. The apo structure of indica rice AKR4C14 was also determined in the absence of the cofactor, revealing the stabilized open conformation. This is the first crystal structure in AKR subfamily 4C from rice to be observed in the apo form (without bound NADP+). The refined AKR4C14 structure reveals a stabilized open conformation of loop B, suggesting the initial phase prior to cofactor binding. Based on the X‐ray crystal structure, the substrate‐ and cofactor‐binding pockets of AKR4C14 are formed by loops A, B, C and β1α1. Moreover, the residues Ser211 and Asn220 on loop B are proposed as the hinge residues that are responsible for conformational alteration while the cofactor binds. The open conformation of loop B is proposed to involve Phe216 pointing out from the cofactor‐binding site and the opening of the safety belt. Structural comparison with other AKRs in subfamily 4C emphasizes the role of the substrate‐channel wall, consisting of Trp24, Trp115, Tyr206, Phe216, Leu291 and Phe295, in substrate discrimination. In particular, Leu291 could contribute greatly to substrate selectivity, explaining the preference of AKR4C14 for its straight‐chain aldehyde substrate.
中文翻译:

AKR4C14的结构功能研究,这是泰国茉莉花米(Oryza sativa L. ssp。indica cv。KDML105)的醛基酮还原酶。
醛酮还原酶(AKR)是NADPH / NADP +依赖性的氧化还原酶,可将醛/酮代谢为相应的醇。水稻中的AKR4C14代谢丙二醛(MDA)的效率比拟南芥酶AKR4C8和AKR4C9高得多,尽管它们具有60%以上的氨基酸序列同一性。这项研究证实了水稻AKR4C14在甲基乙二醛和MDA的解毒中的作用,并证明了转基因拟南芥中两种醛的内源性含量异位表达的AKR4C14明显低于其野生型水平。还在不存在辅因子的情况下测定了rice稻AKR4C14的脱辅基结构,揭示了稳定的开放构象。这是稻的AKR亚家族4C中第一个以apo形式观察到的晶体结构(无结合NADP +)。精制的AKR4C14结构揭示了环B的稳定开放构象,表明辅因子结合之前的初始阶段。基于X射线晶体结构,AKR4C14的底物和辅因子结合袋由环A,B,C和β1α1形成。此外,提出了环B上的残基Ser211和Asn220作为铰链残基,其在辅因子结合时负责构象改变。提出环B的开放构型涉及Phe216从辅因子结合位点和安全带的开口中指出。在亚家族4C中与其他AKR的结构比较强调了底物通道壁(由Trp24,Trp115,Tyr206,Phe216,Leu291和Phe295组成)在底物识别中的作用。特别是Leu291可以大大提高底物的选择性,
更新日期:2020-05-01
中文翻译:

AKR4C14的结构功能研究,这是泰国茉莉花米(Oryza sativa L. ssp。indica cv。KDML105)的醛基酮还原酶。
醛酮还原酶(AKR)是NADPH / NADP +依赖性的氧化还原酶,可将醛/酮代谢为相应的醇。水稻中的AKR4C14代谢丙二醛(MDA)的效率比拟南芥酶AKR4C8和AKR4C9高得多,尽管它们具有60%以上的氨基酸序列同一性。这项研究证实了水稻AKR4C14在甲基乙二醛和MDA的解毒中的作用,并证明了转基因拟南芥中两种醛的内源性含量异位表达的AKR4C14明显低于其野生型水平。还在不存在辅因子的情况下测定了rice稻AKR4C14的脱辅基结构,揭示了稳定的开放构象。这是稻的AKR亚家族4C中第一个以apo形式观察到的晶体结构(无结合NADP +)。精制的AKR4C14结构揭示了环B的稳定开放构象,表明辅因子结合之前的初始阶段。基于X射线晶体结构,AKR4C14的底物和辅因子结合袋由环A,B,C和β1α1形成。此外,提出了环B上的残基Ser211和Asn220作为铰链残基,其在辅因子结合时负责构象改变。提出环B的开放构型涉及Phe216从辅因子结合位点和安全带的开口中指出。在亚家族4C中与其他AKR的结构比较强调了底物通道壁(由Trp24,Trp115,Tyr206,Phe216,Leu291和Phe295组成)在底物识别中的作用。特别是Leu291可以大大提高底物的选择性,



























 京公网安备 11010802027423号
京公网安备 11010802027423号